

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0180a.6
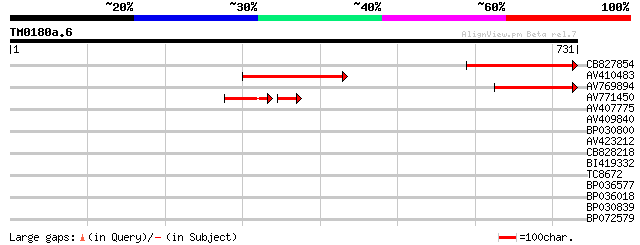
(731 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CB827854 286 1e-77
AV410483 144 4e-35
AV769894 114 4e-26
AV771450 55 3e-12
AV407775 31 0.80
AV409840 30 1.1
BP030800 28 5.2
AV423212 28 5.2
CB828218 28 5.2
BI419332 28 5.2
TC8672 similar to UP|Q8SMH8 (Q8SMH8) RNA-binding protein, partia... 28 6.8
BP036577 28 6.8
BP036018 28 6.8
BP030839 28 6.8
BP072579 27 8.9
>CB827854
Length = 564
Score = 286 bits (731), Expect = 1e-77
Identities = 142/142 (100%), Positives = 142/142 (100%)
Frame = +2
Query: 590 AILMLWRRHFRKGRSLEAFIVTPPPNRNPVEQLLTLQEAITQFESLIQAGNIVLLKVRAL 649
AILMLWRRHFRKGRSLEAFIVTPPPNRNPVEQLLTLQEAITQFESLIQAGNIVLLKVRAL
Sbjct: 2 AILMLWRRHFRKGRSLEAFIVTPPPNRNPVEQLLTLQEAITQFESLIQAGNIVLLKVRAL 181
Query: 650 LLALLPQATEKVALLLVFIAAVFAFVPPKYIIFVIFFEYYTREMPCRKESSDRWVRRMRE 709
LLALLPQATEKVALLLVFIAAVFAFVPPKYIIFVIFFEYYTREMPCRKESSDRWVRRMRE
Sbjct: 182 LLALLPQATEKVALLLVFIAAVFAFVPPKYIIFVIFFEYYTREMPCRKESSDRWVRRMRE 361
Query: 710 WWIRIPAAPVELIKPDECKKRK 731
WWIRIPAAPVELIKPDECKKRK
Sbjct: 362 WWIRIPAAPVELIKPDECKKRK 427
>AV410483
Length = 432
Score = 144 bits (364), Expect = 4e-35
Identities = 72/135 (53%), Positives = 93/135 (68%)
Frame = +2
Query: 301 KPVRYDLGTDLKQVIKPDLTGPLGARLFDKAVMYKSTSVAEPVYFEFPEFKANVRRDYWL 360
KP +YDL DL QV+KP+LTGP G RLFDKAV Y S S++EPV EFPE K + RRDYWL
Sbjct: 2 KPKKYDLSDDLNQVVKPELTGPWGTRLFDKAVFYSSVSLSEPVVLEFPELKGHARRDYWL 181
Query: 361 DISLEILRVHKFIRKYYLKETQKAEVLARAILGIYRYRAVREAFKYFSSHYKTLLSFNLA 420
I EIL VHKFI KY ++ + E L +A+LGI R +A+++ +LL F+L
Sbjct: 182 AIIREILFVHKFISKYRIQGVARDEALWKAVLGILRLQALQDISSAIPIQNDSLLMFSLC 361
Query: 421 EALPRGDMILETLSN 435
+ LP GD+ILETL++
Sbjct: 362 DQLPGGDLILETLAD 406
>AV769894
Length = 467
Score = 114 bits (286), Expect = 4e-26
Identities = 55/107 (51%), Positives = 75/107 (69%)
Frame = -1
Query: 625 LQEAITQFESLIQAGNIVLLKVRALLLALLPQATEKVALLLVFIAAVFAFVPPKYIIFVI 684
+Q A++Q E LIQ GNI+LLK+R LLL++ PQATE +A L+ A V A +P KY++ ++
Sbjct: 467 VQNAVSQAEQLIQDGNIILLKLRGLLLSIFPQATEMLAFALLSAALVLALLPCKYLVLLV 288
Query: 685 FFEYYTREMPCRKESSDRWVRRMREWWIRIPAAPVELIKPDECKKRK 731
F +T P RK S++RW RR+REWW IPAAPV L + E KK+K
Sbjct: 287 FLAVFTMYSPPRKASTERWNRRLREWWFSIPAAPVTLERDKEEKKKK 147
>AV771450
Length = 436
Score = 55.5 bits (132), Expect(2) = 3e-12
Identities = 29/61 (47%), Positives = 41/61 (66%)
Frame = +2
Query: 278 WPGRLTLTNYALYFESLGVGVHEKPVRYDLGTDLKQVIKPDLTGPLGARLFDKAVMYKST 337
WPG+LTLT+ A+YFE+ G+ +++ +R DL D QV+K + GP G+ LFD AV S
Sbjct: 32 WPGKLTLTDKAIYFEAAGLLGNKRAMRLDLTHDGLQVVKAKV-GPFGSALFDSAVSISSG 208
Query: 338 S 338
S
Sbjct: 209 S 211
Score = 33.1 bits (74), Expect(2) = 3e-12
Identities = 13/31 (41%), Positives = 20/31 (63%)
Frame = +3
Query: 346 EFPEFKANVRRDYWLDISLEILRVHKFIRKY 376
EF + +RRD W + E++ +HKFIR+Y
Sbjct: 234 EFIDLGGEMRRDVWHALISEVISLHKFIREY 326
>AV407775
Length = 426
Score = 30.8 bits (68), Expect = 0.80
Identities = 13/29 (44%), Positives = 19/29 (64%), Gaps = 1/29 (3%)
Frame = +3
Query: 345 FEFPEFKANVRRDYWLDISLEILR-VHKF 372
F FP F R WL++S+E+LR +H+F
Sbjct: 69 FRFPRFHFQTRASSWLNLSIELLRHLHEF 155
>AV409840
Length = 417
Score = 30.4 bits (67), Expect = 1.1
Identities = 14/38 (36%), Positives = 20/38 (51%)
Frame = +3
Query: 123 DKDFRRLTYDMMLAWETPSVHTDETPPPSSKDEHGGDD 160
D+ RL + + P HTD++PPPSS + DD
Sbjct: 201 DESMNRLNLLLKDSKPPPRDHTDDSPPPSSSGDKVSDD 314
>BP030800
Length = 516
Score = 28.1 bits (61), Expect = 5.2
Identities = 19/57 (33%), Positives = 26/57 (45%)
Frame = -3
Query: 87 ELPLGVKELLTYARNLLEFCSYKALHKLSKSSDYLADKDFRRLTYDMMLAWETPSVH 143
++ LG+ L Y L E CS +HK KS++ L D D D LA P +
Sbjct: 256 KIALGIARALEY---LHEVCSPSVVHKNIKSANILLDADINPHLSDSGLASYIPGAN 95
>AV423212
Length = 459
Score = 28.1 bits (61), Expect = 5.2
Identities = 14/51 (27%), Positives = 24/51 (46%)
Frame = +3
Query: 560 YKSTAFLLLTSYMILRGWIHYLLPSIFVVVAILMLWRRHFRKGRSLEAFIV 610
+K + LLT+ MI+ W H L ++ + L W + K AF++
Sbjct: 96 FKIIKYFLLTAMMIVERWDHTFLATVSKSMLTLRRWLKKMMKKVPQAAFLL 248
>CB828218
Length = 502
Score = 28.1 bits (61), Expect = 5.2
Identities = 11/34 (32%), Positives = 19/34 (55%), Gaps = 2/34 (5%)
Frame = -3
Query: 692 EMPCRKESSDRWVRRMREW--WIRIPAAPVELIK 723
+M R+ S RW + +W WI +P+ E++K
Sbjct: 359 KMKMRRSSRQRWSPEVNQWWEWILLPSIRTEVVK 258
>BI419332
Length = 471
Score = 28.1 bits (61), Expect = 5.2
Identities = 22/77 (28%), Positives = 38/77 (48%), Gaps = 4/77 (5%)
Frame = +2
Query: 48 AVNSPKPI--PLLSPLANSVVVRSSKILGISTE--ELQHAFDSELPLGVKELLTYARNLL 103
A NSP + PL+ L N++ ++ + +S++ + QH ++ KELL R L
Sbjct: 56 ASNSPTDMANPLVLHLTNNLRLQQPMQIYLSSQVQKPQHRWEGRPLFNPKELLCPKRRLR 235
Query: 104 EFCSYKALHKLSKSSDY 120
+FC A + + DY
Sbjct: 236 QFCWAAASN*VCSREDY 286
>TC8672 similar to UP|Q8SMH8 (Q8SMH8) RNA-binding protein, partial (26%)
Length = 665
Score = 27.7 bits (60), Expect = 6.8
Identities = 16/51 (31%), Positives = 24/51 (46%), Gaps = 2/51 (3%)
Frame = +2
Query: 655 PQATEKVALLLVFIAAVF--AFVPPKYIIFVIFFEYYTREMPCRKESSDRW 703
P +T V LLL+ + +F F + + FF Y PC + SS +W
Sbjct: 20 PLSTTTVLLLLLLLCTLF*TLFF---HFFSLTFFNYPCLSQPCLRSSSSQW 163
>BP036577
Length = 561
Score = 27.7 bits (60), Expect = 6.8
Identities = 19/60 (31%), Positives = 27/60 (44%)
Frame = +2
Query: 575 RGWIHYLLPSIFVVVAILMLWRRHFRKGRSLEAFIVTPPPNRNPVEQLLTLQEAITQFES 634
RG + P I ++V I + RR + RS + F+ PP R+P L T T S
Sbjct: 227 RGDLRLTSPHILLLVTITITRRR---RRRSKQRFLAPPPLYRSPPVNLTTTTTTTTHQSS 397
>BP036018
Length = 488
Score = 27.7 bits (60), Expect = 6.8
Identities = 16/52 (30%), Positives = 23/52 (43%)
Frame = +1
Query: 395 YRYRAVREAFKYFSSHYKTLLSFNLAEALPRGDMILETLSNSLTNLTVASSK 446
+R R YF S +L L LPRG + LSN L N ++ ++
Sbjct: 322 HRLARNRAKLGYFGSQPTSLRVLELFNLLPRGPSVTVNLSNFLLNFPLSLTR 477
>BP030839
Length = 379
Score = 27.7 bits (60), Expect = 6.8
Identities = 15/38 (39%), Positives = 22/38 (57%), Gaps = 5/38 (13%)
Frame = +3
Query: 569 TSYMILRGWIHYLLPSIFVVVAI-----LMLWRRHFRK 601
T+ +IL +HY P I +VVA ++L RRH R+
Sbjct: 192 TTLLILHPQLHYRAPPIIIVVATAA*FRIILRRRHCRR 305
>BP072579
Length = 430
Score = 27.3 bits (59), Expect = 8.9
Identities = 13/28 (46%), Positives = 17/28 (60%)
Frame = +2
Query: 591 ILMLWRRHFRKGRSLEAFIVTPPPNRNP 618
IL R H R+ +SL+ + V P P RNP
Sbjct: 170 ILFRRRHHHRRLKSLKLWAVRPSPLRNP 253
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.135 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,626,706
Number of Sequences: 28460
Number of extensions: 157681
Number of successful extensions: 968
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 953
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 965
length of query: 731
length of database: 4,897,600
effective HSP length: 97
effective length of query: 634
effective length of database: 2,136,980
effective search space: 1354845320
effective search space used: 1354845320
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0180a.6


