

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0177.14
(805 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
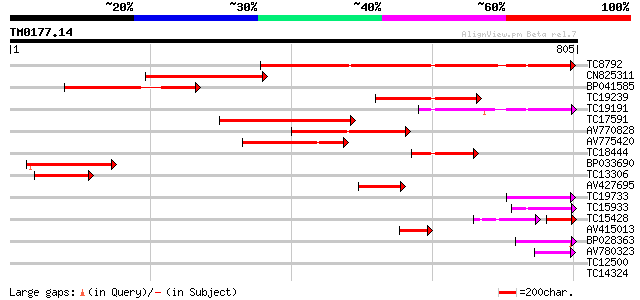
Score E
Sequences producing significant alignments: (bits) Value
TC8792 similar to GB|AAM00218.1|19879972|AF362990 beta-D-xylosid... 388 e-108
CN825311 361 e-100
BP041585 222 2e-58
TC19239 homologue to UP|Q9ZU04 (Q9ZU04) Beta-glucosidase (Fragm... 186 2e-47
TC19191 similar to UP|BAC98299 (BAC98299) LEXYL2 protein (Fragme... 177 8e-45
TC17591 similar to UP|Q9FLG1 (Q9FLG1) Beta-xylosidase, partial (... 175 3e-44
AV770828 157 5e-39
AV775420 139 2e-33
TC18444 similar to UP|Q8W011 (Q8W011) Beta-D-xylosidase, partial... 134 6e-32
BP033690 125 2e-29
TC13306 similar to UP|Q9FGY1 (Q9FGY1) Xylosidase, partial (12%) 101 4e-22
AV427695 82 3e-16
TC19733 weakly similar to UP|Q94KD8 (Q94KD8) At1g02640/T14P4_11,... 76 2e-14
TC15933 weakly similar to UP|BAC98299 (BAC98299) LEXYL2 protein ... 73 2e-13
TC15428 45 1e-11
AV415013 62 5e-10
BP028363 61 8e-10
AV780323 48 7e-06
TC12500 similar to GB|AAH45354.1|28279536|BC045354 FNBP4 protein... 36 0.021
TC14324 weakly similar to UP|Q8LEE9 (Q8LEE9) Arabinogalactan pro... 36 0.028
>TC8792 similar to GB|AAM00218.1|19879972|AF362990 beta-D-xylosidase
{Prunus persica;} , partial (90%)
Length = 1636
Score = 388 bits (996), Expect = e-108
Identities = 204/451 (45%), Positives = 283/451 (62%), Gaps = 3/451 (0%)
Frame = +3
Query: 356 HTASAVEQGKVKEEDIDRALLNLFSVQLRLGLFDGDPRTGKYGKLGPHDVCTSEHKTLAL 415
HT +A++ G + E D++ AL NL +VQ+RLG+FDG+P YG LGP DVCT H LAL
Sbjct: 12 HTDAAIKNGLITENDLNLALANLITVQMRLGMFDGEPSAHPYGNLGPRDVCTPAHNELAL 191
Query: 416 EAARQGIVLLKNDKKFLPLNRNYGSSLAVIGPMA-VTNKLGGGYSGIPCSPKSLYEGLAE 474
EAARQGIVLL+N LPL+ ++ VIGP + VT + G Y+G+ C + +G+A
Sbjct: 192 EAARQGIVLLENKGNALPLSPTRHRTVGVIGPNSDVTVTMIGNYAGVACGYTTPLQGIAR 371
Query: 475 YAKKISYASGCSDISCNSDGGFAEAIETARQADFVVIVAGIDTTLETEDHDRVSLLLPGK 534
Y K I A GC D+SC F A ARQ+D V+V G+D ++E E DRV LLLPG
Sbjct: 372 YVKTIHQA-GCRDVSCRGTELFGNAEIVARQSDATVVVVGLDQSIEAEFRDRVGLLLPGH 548
Query: 535 QMDLVSSIAAASKNPVILVLTGGGPLDVSFAERNQLIPSILWVGYPGEAGGKALAEIIFG 594
Q +LVS +A A++ PV+LV+ GGP+DV FA+ + I +ILWVGYPG+AGG A+A++IFG
Sbjct: 549 QQELVSRVARAARGPVVLVIMSGGPVDVQFAKDDPKISAILWVGYPGQAGGTAIADVIFG 728
Query: 595 ESNPVGFNAAGRLPMTWYPESF-TNVPMNDMSMRADPSRGYPGRTYRFYTGSRVYGFGHG 653
+NP GRLP TWYP+S+ +PM +M MR +P+ GYPGRTYRFY G V+ FGHG
Sbjct: 729 RTNP-----GGRLPNTWYPQSYVAKLPMTNMDMRPNPASGYPGRTYRFYKGPVVFPFGHG 893
Query: 654 LSYSGFSYKFLSAPSKVSLSRITKGSLRKS-LSDQAEKEVYGVDYVQVDELLSCNSLSFP 712
LSYS F++ AP +VS+ + + R S +S +A K + C +L
Sbjct: 894 LSYSRFTHTLAVAPEQVSVPIASLQAFRNSTMSSKAVKVSHA----------KCGALKMG 1043
Query: 713 VHISVTNLGDLDGSHVVMLFSKWPKVVEGSPQTQLVGFSRVHTVSSKSIETSILVDPCEH 772
H+ V N G +DG+H +++FS P + S QLV F + + + ++ V C+H
Sbjct: 1044FHVDVKNEGSMDGTHTLLIFST-PPPGKWSASKQLVSFHKTYVPAGSKQRVNVGVHVCKH 1220
Query: 773 LSFADEQGKRVFPLGNHVLSVGDVEHTVSIE 803
LS DE G R P+G+H L +GDV+HT+S++
Sbjct: 1221LSVVDENGIRRIPMGDHELHIGDVKHTISVQ 1313
>CN825311
Length = 522
Score = 361 bits (926), Expect = e-100
Identities = 172/173 (99%), Positives = 172/173 (99%)
Frame = +2
Query: 193 KNVFGEKRALSDYGGGSGNGSFDSDSDDGDDGDSLMVSACCKHFTAYDLEKWGQFARYNF 252
KNVFGEKRALSDYGGGSGNGSFDSDSDDGDDGDSLMVSACCKHFTAYDLEKWGQFARYNF
Sbjct: 2 KNVFGEKRALSDYGGGSGNGSFDSDSDDGDDGDSLMVSACCKHFTAYDLEKWGQFARYNF 181
Query: 253 NAVVSQQDLEDTYQPPFRGCVQQGKASCLMCSYNEVNGVPACASEDLLGVARNNWGFKGY 312
NAVVSQQDLEDTYQPPFRGCVQQGKASCLMCSYNEVNGVPACASEDLLGVARNNWGFKGY
Sbjct: 182 NAVVSQQDLEDTYQPPFRGCVQQGKASCLMCSYNEVNGVPACASEDLLGVARNNWGFKGY 361
Query: 313 ITSDCDAVATVFEYQGYVKSAEDAVAEVLKAGTDINCGTYMLRHTASAVEQGK 365
ITSDCDAVATVFEYQ YVKSAEDAVAEVLKAGTDINCGTYMLRHTASAVEQGK
Sbjct: 362 ITSDCDAVATVFEYQVYVKSAEDAVAEVLKAGTDINCGTYMLRHTASAVEQGK 520
>BP041585
Length = 503
Score = 222 bits (565), Expect = 2e-58
Identities = 105/193 (54%), Positives = 131/193 (67%)
Frame = +1
Query: 78 PAYQWWSESLHGIAINGPGVSFDGAVSAATDFPQVIVSAASFNRTLWFLIASAVAVEARA 137
P+Y+WWSE+LHG++ GPG F+ V AT FP VI+SAASFN TLW+ + V+ EARA
Sbjct: 1 PSYEWWSEALHGVSNLGPGTRFNRTVPGATSFPAVILSAASFNATLWYNMGQVVSTEARA 180
Query: 138 MFNVGQAGLTFWAPNVNVFRDPRWGRGQETPGEDPMVASAYAVDYVRGLQGAGGVKNVFG 197
M+NV AGLTFW+PNVNVFRDPRWGRGQETPGEDP+V S YAV YVRGLQ
Sbjct: 181 MYNVDLAGLTFWSPNVNVFRDPRWGRGQETPGEDPLVVSRYAVKYVRGLQ---------- 330
Query: 198 EKRALSDYGGGSGNGSFDSDSDDGDDGDSLMVSACCKHFTAYDLEKWGQFARYNFNAVVS 257
D + ++ D L VS+CCKH+TAYD++ W R++F+A V+
Sbjct: 331 -----------------DVEDEESIKADRLKVSSCCKHYTAYDVDNWKGVDRFHFDAKVT 459
Query: 258 QQDLEDTYQPPFR 270
+QDLEDTYQPPF+
Sbjct: 460 KQDLEDTYQPPFK 498
>TC19239 homologue to UP|Q9ZU04 (Q9ZU04) Beta-glucosidase (Fragment) ,
partial (69%)
Length = 453
Score = 186 bits (471), Expect = 2e-47
Identities = 91/152 (59%), Positives = 112/152 (72%), Gaps = 1/152 (0%)
Frame = +1
Query: 520 ETEDHDRVSLLLPGKQMDLVSSIAAASKNPVILVLTGGGPLDVSFAERNQLIPSILWVGY 579
+ E DRV+++LPG+Q LVS +A AS+ PVILV+ GG +DVSFA+ N I SILWVGY
Sbjct: 1 QAESLDRVNIVLPGQQQLLVSEVANASRGPVILVIMSGGGMDVSFAKANDKITSILWVGY 180
Query: 580 PGEAGGKALAEIIFGESNPVGFNAAGRLPMTWYPESFT-NVPMNDMSMRADPSRGYPGRT 638
PGEAGG A+A++IFG NP +GRLPMTWYPES+ NVPM +M+MRADPS GYPGRT
Sbjct: 181 PGEAGGAAIADVIFGFYNP-----SGRLPMTWYPESYVENVPMTNMNMRADPSTGYPGRT 345
Query: 639 YRFYTGSRVYGFGHGLSYSGFSYKFLSAPSKV 670
YRFY G V+ FG G+ YS +K + AP V
Sbjct: 346 YRFYKGETVFSFGDGIGYSNVEHKIVKAPQLV 441
>TC19191 similar to UP|BAC98299 (BAC98299) LEXYL2 protein (Fragment),
partial (31%)
Length = 770
Score = 177 bits (448), Expect = 8e-45
Identities = 97/229 (42%), Positives = 126/229 (54%), Gaps = 5/229 (2%)
Frame = +2
Query: 581 GEAGGKALAEIIFGESNPVGFNAAGRLPMTWYPESFTN-VPMNDMSMRADPSRGYPGRTY 639
GEAGG A+A++IFG NP +GRLPMTWYP+S+ + VPM +M+MR DP+ GYPGRTY
Sbjct: 2 GEAGGAAIADVIFGFHNP-----SGRLPMTWYPQSYVDKVPMTNMNMRPDPTTGYPGRTY 166
Query: 640 RFYTGSRVYGFGHGLSYSGFSYKFLSAPSKVSL----SRITKGSLRKSLSDQAEKEVYGV 695
RFY G V+ FG GLSYS +K + AP VS+ + KSL E
Sbjct: 167 RFYKGETVFSFGDGLSYSTIEHKLVKAPQLVSIPLAEDHVCHSLKCKSLDVAGE------ 328
Query: 696 DYVQVDELLSCNSLSFPVHISVTNLGDLDGSHVVMLFSKWPKVVEGSPQTQLVGFSRVHT 755
C +L F +H+S+ N G + H V LFS P V +P LVGF +VH
Sbjct: 329 ---------HCENLEFEIHLSIKNKGRMSSDHTVFLFSS-PPAVHNAPHKHLVGFEKVHL 478
Query: 756 VSSKSIETSILVDPCEHLSFADEQGKRVFPLGNHVLSVGDVEHTVSIEI 804
+ VD C+ LS DE G R LG H+L VGD++H + + I
Sbjct: 479 EGNTEALVKFKVDVCKDLSVVDELGNRKVALGQHLLHVGDLKHALGVMI 625
>TC17591 similar to UP|Q9FLG1 (Q9FLG1) Beta-xylosidase, partial (24%)
Length = 582
Score = 175 bits (443), Expect = 3e-44
Identities = 95/193 (49%), Positives = 121/193 (62%), Gaps = 1/193 (0%)
Frame = +3
Query: 299 LLGVARNNWGFKGYITSDCDAVATVFEYQGYVKSAEDAVAEVLKAGTDINCGTYMLRHTA 358
L GV R W GYI SDCD+V +F+ Q Y K+ E+A A+ + AG D++CG+Y+ ++T
Sbjct: 3 LKGVIRGQWKLNGYIVSDCDSVEVLFKDQHYTKTPEEAAAKSILAGLDLDCGSYLGKYTE 182
Query: 359 SAVEQGKVKEEDIDRALLNLFSVQLRLGLFDGDPRTGKYGKLGPHDVCTSEHKTLALEAA 418
AV+QG + E I+ A+ N F+ +RLG FDG+P T YG LGP DVCTSE++ LA EAA
Sbjct: 183 GAVKQGLLDEASINNAISNNFATLMRLGFFDGNPSTLPYGGLGPKDVCTSENQELAREAA 362
Query: 419 RQGIVLLKNDKKFLPLNRNYGSSLAVIGPMA-VTNKLGGGYSGIPCSPKSLYEGLAEYAK 477
RQGIVLLKN LPL+ SLAVIGP A T + G GIPC S +GL
Sbjct: 363 RQGIVLLKNSPGSLPLSAKSIKSLAVIGPNANATRVMIGN*EGIPCKYISPLQGLTALV- 539
Query: 478 KISYASGCSDISC 490
SY GC D+ C
Sbjct: 540 PTSYVPGCPDVHC 578
>AV770828
Length = 527
Score = 157 bits (398), Expect = 5e-39
Identities = 86/169 (50%), Positives = 112/169 (65%), Gaps = 1/169 (0%)
Frame = +1
Query: 401 GPHDVCTSEHKTLALEAARQGIVLLKNDKKFLPLNRNYGSSLAVIGPMA-VTNKLGGGYS 459
GP DVCT H+ LALEAARQGIVLLKN LPL+ +LA+IGP + T + G Y+
Sbjct: 22 GPRDVCTQPHQELALEAARQGIVLLKNSGPSLPLSTRRHRTLAIIGPNSNATVTMIGNYA 201
Query: 460 GIPCSPKSLYEGLAEYAKKISYASGCSDISCNSDGGFAEAIETARQADFVVIVAGIDTTL 519
GI S EG+ +YAK I + GC++++ D F A+ ARQAD V+V G+D ++
Sbjct: 202 GIGYRYTSP*EGIGKYAKTI-HELGCANVAGTDDKQFGSALNAARQADATVLVMGLDQSI 378
Query: 520 ETEDHDRVSLLLPGKQMDLVSSIAAASKNPVILVLTGGGPLDVSFAERN 568
E E DR LLLPG+Q DLVS +AAASK P ILVL GGP+D++FA+ +
Sbjct: 379 EAETVDRAGLLLPGRQQDLVSKVAAASKGPTILVLMSGGPVDITFAKND 525
>AV775420
Length = 457
Score = 139 bits (350), Expect = 2e-33
Identities = 76/153 (49%), Positives = 97/153 (62%), Gaps = 2/153 (1%)
Frame = +1
Query: 331 KSAEDAVAEVLKAGTDINCGTYMLRHTASAVEQGKVKEEDIDRALLNLFSVQLRLGLFDG 390
K++ VA+VL+AG D+ CG Y+ +H SAV Q KV IDRAL NLFS+++RLGLFDG
Sbjct: 1 KNSRGCVADVLRAGMDVECGDYLTKHGKSAV*QKKVPISQIDRALHNLFSIRIRLGLFDG 180
Query: 391 DPRTGKYGKLGPHDVCTSEHKTLALEAARQGIVLLKNDKKFLPLNRNYGSSLAVIGPMAV 450
+P YG +GP+ VC+ ++ LALEAAR GIVLLK LPL + S+AVIGP A
Sbjct: 181 NPSKLTYGSIGPNQVCSKQNLKLALEAARNGIVLLKKTASILPLPKT-NPSMAVIGPNAN 357
Query: 451 TNKLG--GGYSGIPCSPKSLYEGLAEYAKKISY 481
+ L G Y G PC +L +G YAK Y
Sbjct: 358 ASSLAVLGNYYGRPCKLVTLLQGFQHYAKDTIY 456
>TC18444 similar to UP|Q8W011 (Q8W011) Beta-D-xylosidase, partial (12%)
Length = 387
Score = 134 bits (337), Expect = 6e-32
Identities = 60/95 (63%), Positives = 73/95 (76%)
Frame = +2
Query: 571 IPSILWVGYPGEAGGKALAEIIFGESNPVGFNAAGRLPMTWYPESFTNVPMNDMSMRADP 630
I ILW GYPGE GG ALA++IFG+ NP GRLP+TWYP+ + VPM DM MRADP
Sbjct: 5 IGGILWAGYPGELGGIALAQVIFGDHNP-----GGRLPITWYPKDYIKVPMTDMRMRADP 169
Query: 631 SRGYPGRTYRFYTGSRVYGFGHGLSYSGFSYKFLS 665
+ GYPGRTYRFY G ++Y FG+GLSYS +SY+F+S
Sbjct: 170 ATGYPGRTYRFYKGPKLYEFGYGLSYSKYSYEFIS 274
>BP033690
Length = 546
Score = 125 bits (315), Expect = 2e-29
Identities = 65/130 (50%), Positives = 83/130 (63%), Gaps = 3/130 (2%)
Frame = +2
Query: 25 DFAC---KRPHHSRYKFCDTSLSIPTRAHSLVSLLTLPEKIQQLSNNASSIPRLGIPAYQ 81
+FAC K P S Y FC+ SL + R LV LTL EKI L + A+ + RLGIP Y+
Sbjct: 155 NFACDVAKNPALSGYGFCNRSLPVNARVADLVGRLTLQEKIGNLVSGAAIVSRLGIPKYE 334
Query: 82 WWSESLHGIAINGPGVSFDGAVSAATDFPQVIVSAASFNRTLWFLIASAVAVEARAMFNV 141
WWSE+LHG++ GPG F V A FP I++AASFN +L+ I V+ EARAM+NV
Sbjct: 335 WWSEALHGVSNIGPGTRFSNVVPRAASFPMPILTAASFNTSLFQAIGKVVSTEARAMYNV 514
Query: 142 GQAGLTFWAP 151
G AGLT+ +P
Sbjct: 515 GLAGLTYGSP 544
>TC13306 similar to UP|Q9FGY1 (Q9FGY1) Xylosidase, partial (12%)
Length = 435
Score = 101 bits (252), Expect = 4e-22
Identities = 47/84 (55%), Positives = 59/84 (69%)
Frame = +2
Query: 36 YKFCDTSLSIPTRAHSLVSLLTLPEKIQQLSNNASSIPRLGIPAYQWWSESLHGIAINGP 95
+KFC T++ I R L+ LTL EKI+ + NNA +PRLGI Y+WWSE+LHG++ GP
Sbjct: 182 FKFCRTNVPIHVRVQDLIGRLTLAEKIRLVVNNAIEVPRLGIQGYEWWSEALHGVSNVGP 361
Query: 96 GVSFDGAVSAATDFPQVIVSAASF 119
G F GA AAT FPQVI +AASF
Sbjct: 362 GTKFGGAFPAATSFPQVITTAASF 433
>AV427695
Length = 202
Score = 82.0 bits (201), Expect = 3e-16
Identities = 39/67 (58%), Positives = 48/67 (71%)
Frame = +2
Query: 496 FAEAIETARQADFVVIVAGIDTTLETEDHDRVSLLLPGKQMDLVSSIAAASKNPVILVLT 555
F A++ AR AD V+V G+D ++E E DRV LLLPG Q DLVS +AAAS+ P ILVL
Sbjct: 2 FGPALDAARHADATVLVMGLDQSIEAETRDRVGLLLPGHQQDLVSKVAAASRGPTILVLM 181
Query: 556 GGGPLDV 562
GGPLD+
Sbjct: 182 SGGPLDI 202
>TC19733 weakly similar to UP|Q94KD8 (Q94KD8) At1g02640/T14P4_11, partial
(15%)
Length = 598
Score = 75.9 bits (185), Expect = 2e-14
Identities = 37/99 (37%), Positives = 56/99 (56%), Gaps = 1/99 (1%)
Frame = +2
Query: 706 CNSLSFPVHISVTNLGDLDGSHVVMLFSKWPKV-VEGSPQTQLVGFSRVHTVSSKSIETS 764
C LS +H++V N+G DG+H +++FS P + Q QLV F +VH +
Sbjct: 89 CGKLSIALHVNVKNVGSRDGTHTLLVFSAPPDGGAHWAVQKQLVAFEKVHVPAKSQQRVQ 268
Query: 765 ILVDPCEHLSFADEQGKRVFPLGNHVLSVGDVEHTVSIE 803
I + C+ LS D G R P+G H L +GDV+H+VS++
Sbjct: 269 INIHVCKLLSVVDRSGIRRIPMGEHSLHIGDVKHSVSLQ 385
>TC15933 weakly similar to UP|BAC98299 (BAC98299) LEXYL2 protein (Fragment),
partial (14%)
Length = 453
Score = 73.2 bits (178), Expect = 2e-13
Identities = 34/92 (36%), Positives = 52/92 (55%)
Frame = +1
Query: 713 VHISVTNLGDLDGSHVVMLFSKWPKVVEGSPQTQLVGFSRVHTVSSKSIETSILVDPCEH 772
+H+ V N+G + SH V+LF P V +PQ L+G +VH + +VD C+
Sbjct: 1 IHLRVKNMGKMTSSHTVLLFFT-PPAVHNAPQKHLLGIEKVHLAGKSEAQVRFMVDVCKD 177
Query: 773 LSFADEQGKRVFPLGNHVLSVGDVEHTVSIEI 804
LS DE R PLG H+L VG++++ +S+ I
Sbjct: 178 LSVVDELAHRRVPLGQHLLHVGNLKYPLSVRI 273
>TC15428
Length = 629
Score = 44.7 bits (104), Expect(2) = 1e-11
Identities = 19/43 (44%), Positives = 29/43 (67%)
Frame = +2
Query: 762 ETSILVDPCEHLSFADEQGKRVFPLGNHVLSVGDVEHTVSIEI 804
E + PCEHLS A+E G +V P G+++L VGD E+ ++I +
Sbjct: 302 EVGFELSPCEHLSIANEGGVKVIPEGSYLLHVGDQEYPMNISV 430
Score = 42.0 bits (97), Expect(2) = 1e-11
Identities = 31/97 (31%), Positives = 48/97 (48%), Gaps = 2/97 (2%)
Frame = +1
Query: 659 FSYKFLSAPSKVSLSRITKGSLRKSLSDQAEKEVYGVDYVQVDEL--LSCNSLSFPVHIS 716
+SY+F+S + I + S ++ + +E + Y V EL +C S+S V +
Sbjct: 4 YSYEFVSVTQ--NNLHINQSSTHLTVDENSET----IRYKLVSELGEETCQSMSVSVTLG 165
Query: 717 VTNLGDLDGSHVVMLFSKWPKVVEGSPQTQLVGFSRV 753
V N G + G H V+LF + K G+P LVGF V
Sbjct: 166 VKNDGSMAGKHPVLLFMRPGKQRNGNPLKPLVGFPSV 276
>AV415013
Length = 154
Score = 61.6 bits (148), Expect = 5e-10
Identities = 26/47 (55%), Positives = 35/47 (74%)
Frame = +2
Query: 554 LTGGGPLDVSFAERNQLIPSILWVGYPGEAGGKALAEIIFGESNPVG 600
+ GP+D+SF + + I ILWVGYPG+AGG A+A++IFGE NP G
Sbjct: 2 IMAAGPIDISFTKSIRNIGGILWVGYPGQAGGDAIAQVIFGEYNPGG 142
>BP028363
Length = 508
Score = 60.8 bits (146), Expect = 8e-10
Identities = 34/93 (36%), Positives = 51/93 (54%), Gaps = 7/93 (7%)
Frame = -3
Query: 719 NLGDLDGSHVVMLFSKWP--KVVEGSPQTQLVGFSRVHTVSSKSIETSILVDPCEHLSFA 776
N G +GSHVV++F + P ++V G+P QL+GF R H + + ++ +D C+ LS
Sbjct: 503 NTGPYNGSHVVLVFWEPPTSELVTGAPIKQLIGFERAHVMVGMTEFVTLEIDICQLLSNV 324
Query: 777 DEQGKRVFPLGNHVLSVG-----DVEHTVSIEI 804
D GKR +G H L VG V H + I +
Sbjct: 323 DSDGKRKLIIGKHTLLVGSSSETQVRHQIDISL 225
>AV780323
Length = 544
Score = 47.8 bits (112), Expect = 7e-06
Identities = 20/58 (34%), Positives = 32/58 (54%)
Frame = -3
Query: 746 QLVGFSRVHTVSSKSIETSILVDPCEHLSFADEQGKRVFPLGNHVLSVGDVEHTVSIE 803
QLV F +VH + + + C+ LS D G R P+G H +GD+EH++S++
Sbjct: 536 QLVAFEKVHIPAKAQRRVRVNIHVCKLLSVVDRSGIRRIPMGEHRFQIGDIEHSLSLQ 363
>TC12500 similar to GB|AAH45354.1|28279536|BC045354 FNBP4 protein {Danio
rerio;} , partial (5%)
Length = 678
Score = 36.2 bits (82), Expect = 0.021
Identities = 21/58 (36%), Positives = 30/58 (51%)
Frame = -2
Query: 163 RGQETPGEDPMVASAYAVDYVRGLQGAGGVKNVFGEKRALSDYGGGSGNGSFDSDSDD 220
+G+ET D A+ ++G +G GG G RALSD+GG G GS ++D
Sbjct: 179 QGRETSPTDLETFEIIALPAIKGAEGGGG-----GGGRALSDFGGVEGGGSGALSAED 21
>TC14324 weakly similar to UP|Q8LEE9 (Q8LEE9) Arabinogalactan protein-like,
partial (63%)
Length = 1172
Score = 35.8 bits (81), Expect = 0.028
Identities = 19/59 (32%), Positives = 30/59 (50%)
Frame = +3
Query: 104 SAATDFPQVIVSAASFNRTLWFLIASAVAVEARAMFNVGQAGLTFWAPNVNVFRDPRWG 162
S+ TD +++ A + + L + VA + A AGLTF+APN N F + + G
Sbjct: 201 SSPTDIIRILKKAGGYTTLIRLLQTTQVATQINAQLINSNAGLTFFAPNDNAFSNLKPG 377
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.136 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,726,957
Number of Sequences: 28460
Number of extensions: 186812
Number of successful extensions: 1023
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 991
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1001
length of query: 805
length of database: 4,897,600
effective HSP length: 98
effective length of query: 707
effective length of database: 2,108,520
effective search space: 1490723640
effective search space used: 1490723640
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0177.14


