

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
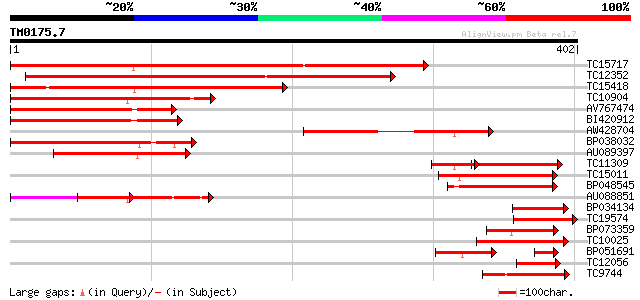
Query= TM0175.7
(402 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC15717 weakly similar to UP|Q8GT65 (Q8GT65) Serpin-like protein... 349 4e-97
TC12352 weakly similar to UP|Q8GT65 (Q8GT65) Serpin-like protein... 324 2e-89
TC15418 weakly similar to UP|Q8GT65 (Q8GT65) Serpin-like protein... 213 3e-56
TC10904 weakly similar to UP|Q62258 (Q62258) Spi2 proteinase inh... 173 4e-44
AV767474 171 2e-43
BI420912 154 2e-38
AW428704 119 7e-28
BP038032 112 1e-25
AU089397 107 4e-24
TC11309 similar to UP|Q9FUV8 (Q9FUV8) Phloem serpin-1, partial (... 78 2e-20
TC15011 similar to UP|Q8GT65 (Q8GT65) Serpin-like protein (Fragm... 88 2e-18
BP048545 82 2e-16
AU088851 78 2e-15
BP034134 69 1e-12
TC19574 weakly similar to UP|Q8GT65 (Q8GT65) Serpin-like protein... 66 9e-12
BP073359 65 2e-11
TC10025 weakly similar to UP|Q8GT65 (Q8GT65) Serpin-like protein... 65 3e-11
BP051691 49 5e-11
TC12056 weakly similar to UP|ILEU_HUMAN (P30740) Leukocyte elast... 62 1e-10
TC9744 similar to UP|Q40066 (Q40066) Protein zx, partial (7%) 62 2e-10
>TC15717 weakly similar to UP|Q8GT65 (Q8GT65) Serpin-like protein
(Fragment), partial (26%)
Length = 925
Score = 349 bits (896), Expect = 4e-97
Identities = 186/303 (61%), Positives = 221/303 (72%), Gaps = 6/303 (1%)
Frame = +1
Query: 1 MDLQKSI-RCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELF 59
M+LQKSI + Q +VALS LFS E Y +N+VFSP SLH L++MA G+ GSTLDEL
Sbjct: 19 MELQKSISKSQEDVALSFANRLFSTEAYHNENIVFSPLSLHVALAIMAAGAHGSTLDELL 198
Query: 60 SFLRFDSVDHLNTFFSQVLSTVLSDTT-----PSFLLSFVNEMWADKSLALSHSFKQLMT 114
SFLRFDSVDHLNT FSQV+S V SD P+ LSF N MW DKSL+L+HSFKQL+
Sbjct: 199 SFLRFDSVDHLNTIFSQVVSAVFSDNDDAAPPPTHRLSFANGMWVDKSLSLTHSFKQLVA 378
Query: 115 THYKATLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKG 174
THYKATL VDF K DQVC EVN WVEK TNGLI ELL AV K T+LIFANAL+FKG
Sbjct: 379 THYKATLDSVDFWNKADQVCDEVNLWVEKGTNGLIKELLSPGAVDKTTRLIFANALHFKG 558
Query: 175 VWKHTFDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLR 234
W+H F ++ FHLL+GTS+ VP M+++++ Q IR FDGF +L L YKQG D+K
Sbjct: 559 EWEHKFLARYSYSYRFHLLDGTSVVVPLMTNDEE-QLIRVFDGFKILGLPYKQGTDEKRL 735
Query: 235 FSMCIFLPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLK 294
FSM I LP+AKDGL LI K+ASE FL+GKLP+QKV++ F IP+F I F EAS+VLK
Sbjct: 736 FSMYILLPHAKDGLSDLIRKMASEPGFLEGKLPQQKVKLNFFLIPRFDISFAFEASDVLK 915
Query: 295 ELG 297
E G
Sbjct: 916 EFG 924
>TC12352 weakly similar to UP|Q8GT65 (Q8GT65) Serpin-like protein
(Fragment), partial (25%)
Length = 799
Score = 324 bits (830), Expect = 2e-89
Identities = 157/262 (59%), Positives = 207/262 (78%)
Frame = +1
Query: 12 EVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFSFLRFDSVDHLN 71
+V LS T+HLFSKE+YQEKNL++SP SL+A LSV+A GS G T DEL SFLRFDS+D+LN
Sbjct: 16 DVPLSFTQHLFSKEDYQEKNLIYSPLSLYAALSVIAAGSEGRTFDELLSFLRFDSIDNLN 195
Query: 72 TFFSQVLSTVLSDTTPSFLLSFVNEMWADKSLALSHSFKQLMTTHYKATLALVDFRTKGD 131
TFFSQ +S V D + L N ++ D +++LS+ F++L++THY A L +DF +G
Sbjct: 196 TFFSQAISPVFFDNDAASPLQHYNGIFIDTTVSLSYPFRRLLSTHYNANLTSLDFNLRGG 375
Query: 132 QVCREVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKGVWKHTFDPSMTFVDDFH 191
+V E+NS +E++TNG IT+LLP V LT+LIFANAL F+G+WKH FD +T+V F+
Sbjct: 376 KVLHEMNSLIEEDTNGHITQLLPPGTVTNLTRLIFANALCFQGMWKHKFD-GLTYVSPFN 552
Query: 192 LLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLRFSMCIFLPNAKDGLPSL 251
LLNGTS++VPFM++ K TQ++R FDGF +LRL YKQGRD++ RFSMCIFLP+A+DGL +L
Sbjct: 553 LLNGTSVKVPFMTTCKNTQYVRAFDGFKILRLPYKQGRDRQRRFSMCIFLPDAQDGLSAL 732
Query: 252 IEKLASESCFLKGKLPRQKVRV 273
I+KL+SE CFLKGKLPR+KVRV
Sbjct: 733 IQKLSSEPCFLKGKLPRRKVRV 798
>TC15418 weakly similar to UP|Q8GT65 (Q8GT65) Serpin-like protein
(Fragment), partial (44%)
Length = 602
Score = 213 bits (543), Expect = 3e-56
Identities = 112/199 (56%), Positives = 143/199 (71%), Gaps = 2/199 (1%)
Frame = +1
Query: 1 MDLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFS 60
MDL++SI QT+V+L+I K LFSK ++KN VFSP SLH +LS++A GS G TL +L +
Sbjct: 7 MDLRESIETQTDVSLTIAKLLFSKHS-KDKNAVFSPLSLHVVLSILAAGSEGPTLHQLLT 183
Query: 61 FLRFDSVDHLNTFFSQVLSTVLSDTTP--SFLLSFVNEMWADKSLALSHSFKQLMTTHYK 118
FLR +S DHLN+F SQ++S VLSD + L F + +W ++SL L+ SFKQL+T Y
Sbjct: 184 FLRSNSTDHLNSFASQLVSVVLSDASSVGGPRLCFADGVWVEQSLTLNPSFKQLVTADYH 363
Query: 119 ATLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKGVWKH 178
A LA VDF+TK +V EVN+W EKETNGL+ +LLP +V T+LIFANALYFKG W
Sbjct: 364 AALASVDFQTKAVEVANEVNAWAEKETNGLVKDLLPAGSVDASTRLIFANALYFKGAWTE 543
Query: 179 TFDPSMTFVDDFHLLNGTS 197
FD SMT DFHLL+GTS
Sbjct: 544 KFDASMTKDCDFHLLDGTS 600
>TC10904 weakly similar to UP|Q62258 (Q62258) Spi2 proteinase inhibitor,
partial (8%)
Length = 576
Score = 173 bits (439), Expect = 4e-44
Identities = 94/153 (61%), Positives = 111/153 (72%), Gaps = 7/153 (4%)
Frame = -1
Query: 1 MDLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFS 60
M+L+KS +VAL +TKHLFSK +YQ KN+VFSPFSL A LSVMA GS TLDEL S
Sbjct: 453 MELEKSKSKSMDVALGLTKHLFSKADYQGKNIVFSPFSLQAALSVMAAGSKDRTLDELLS 274
Query: 61 FLRFDSVDHLNTFFSQVLSTVL-------SDTTPSFLLSFVNEMWADKSLALSHSFKQLM 113
FLRFDS+D L TFFSQV+ VL +DT S L F N +WAD SL+LSH FKQL+
Sbjct: 273 FLRFDSIDDLTTFFSQVIFPVLISDAAADADTDGSHHLCFANGIWADDSLSLSHRFKQLV 94
Query: 114 TTHYKATLALVDFRTKGDQVCREVNSWVEKETN 146
THYKATL +DF+T +V REVNSW+EKET+
Sbjct: 93 ATHYKATLTALDFQT--TEVHREVNSWIEKETD 1
>AV767474
Length = 380
Score = 171 bits (433), Expect = 2e-43
Identities = 82/118 (69%), Positives = 102/118 (85%)
Frame = +3
Query: 1 MDLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFS 60
MDL++ +RCQT+VAL+ITKHLFSK+EYQ+KNL+FSP SLHA+LS++A GSAGSTLDEL S
Sbjct: 33 MDLRELMRCQTDVALTITKHLFSKQEYQDKNLMFSPLSLHAVLSLLAAGSAGSTLDELLS 212
Query: 61 FLRFDSVDHLNTFFSQVLSTVLSDTTPSFLLSFVNEMWADKSLALSHSFKQLMTTHYK 118
FLRFDS+DHLNTFFSQ++S + P L+FVN MW DKS++LS+SFKQL+ THYK
Sbjct: 213 FLRFDSIDHLNTFFSQLISAAAPSSHP---LTFVNGMWVDKSVSLSNSFKQLLATHYK 377
>BI420912
Length = 508
Score = 154 bits (390), Expect = 2e-38
Identities = 85/123 (69%), Positives = 94/123 (76%), Gaps = 1/123 (0%)
Frame = +2
Query: 1 MDLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFS 60
MDLQKS + LS TKH+ SKEEYQEKNLVFSP SL+A LSVMA G+ G TLDEL S
Sbjct: 146 MDLQKSKSNSIDAVLSFTKHVLSKEEYQEKNLVFSPLSLYAALSVMAAGADGLTLDELLS 325
Query: 61 FLRFDSVDHLNTFFSQVLSTVL-SDTTPSFLLSFVNEMWADKSLALSHSFKQLMTTHYKA 119
FLRFDSVDHL TFFSQ LS VL SD LSF N MW D+SL++ HSFKQL++THYKA
Sbjct: 326 FLRFDSVDHLTTFFSQDLSPVLFSDDH----LSFANGMWGDQSLSIFHSFKQLVSTHYKA 493
Query: 120 TLA 122
TLA
Sbjct: 494 TLA 502
>AW428704
Length = 369
Score = 119 bits (299), Expect = 7e-28
Identities = 73/137 (53%), Positives = 84/137 (61%), Gaps = 2/137 (1%)
Frame = +1
Query: 209 TQFIRTFDGFNVLRLSYKQGRDKKLRFSMCIFLPNAKDGLPSLIEKLASESCFLKGKLPR 268
T+ IR FDGF +L L YKQG D+K FSM I LP+AKDGL LI K+ASE F
Sbjct: 10 TRLIRVFDGFKILGLPYKQGTDEKRLFSMYILLPHAKDGLSDLIRKMASEPGF------- 168
Query: 269 QKVRVRKFRIPKFKICFELEASNVLKELGVVSPFSKSNANFTKMVV--SPLDELCVESIH 326
LEAS+VLKE GVVSPFS+ +A+FTKMV SPLD L VESI
Sbjct: 169 ------------------LEASDVLKEFGVVSPFSQRDADFTKMVKVNSPLDALSVESIF 294
Query: 327 HKASIEVNEEGTEAGVA 343
K I+VNE+GTEA A
Sbjct: 295 QKVFIKVNEQGTEAAAA 345
>BP038032
Length = 437
Score = 112 bits (279), Expect = 1e-25
Identities = 70/142 (49%), Positives = 86/142 (60%), Gaps = 10/142 (7%)
Frame = +1
Query: 1 MDLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELF- 59
MD++KS+RCQT+VALS+TKHLFSKEEYQ+KNL+FSP SLH LSVMA GSAG TL F
Sbjct: 16 MDVEKSMRCQTDVALSVTKHLFSKEEYQQKNLIFSPLSLHVALSVMAGGSAGGTLSRTFL 195
Query: 60 SFLRFDS-VDHLNTFFSQVLSTVLSDTTPSFL----LSFVNEMWADKSLALSHSFKQLMT 114
SF S + N + +L+ TP + L N MW D ++ F L
Sbjct: 196 SFFGIGSHWKNFNNLSNLILNFHRGPWTPVVISPLRLPLANGMWVD---*ITFHFPILPN 366
Query: 115 T----HYKATLALVDFRTKGDQ 132
HYKATLA +DF+ GDQ
Sbjct: 367 NW*PLHYKATLASLDFQKNGDQ 432
>AU089397
Length = 306
Score = 107 bits (267), Expect = 4e-24
Identities = 56/99 (56%), Positives = 70/99 (70%), Gaps = 2/99 (2%)
Frame = +3
Query: 32 LVFSPFSLHAILSVMAVGSAGSTLDELFSFLRFDSVDHLNTFFSQVLSTVLSDTTPSF-- 89
+VFSP SL +LS++A GS GSTLDEL SFL +S DHLN+F SQ++STVLSD P+
Sbjct: 9 IVFSPLSLQVVLSIIAAGSDGSTLDELLSFLGSNSTDHLNSFASQLISTVLSDAAPAGGP 188
Query: 90 LLSFVNEMWADKSLALSHSFKQLMTTHYKATLALVDFRT 128
L F +W +KSL ++HSF Q+M T Y ATL VDF T
Sbjct: 189 RLCFAXXVWVEKSLPVNHSFXQVMNTDYXATLTSVDFCT 305
>TC11309 similar to UP|Q9FUV8 (Q9FUV8) Phloem serpin-1, partial (10%)
Length = 472
Score = 77.8 bits (190), Expect(2) = 2e-20
Identities = 41/66 (62%), Positives = 47/66 (71%), Gaps = 1/66 (1%)
Frame = +3
Query: 328 KASIEVNEEGTEAGVACSMCDVKLC-VSARTGIDFVADHPFLFLIREDLTGTILFIGQVL 386
+ S++++EEGT A A +M S R GIDFVADHPF FLIRED TGTILFIGQVL
Sbjct: 96 RLSLKLDEEGTTAAAATAMLLASRSGPSVRAGIDFVADHPFFFLIREDFTGTILFIGQVL 275
Query: 387 HPEGAA 392
HP AA
Sbjct: 276 HPNVAA 293
Score = 38.1 bits (87), Expect(2) = 2e-20
Identities = 22/37 (59%), Positives = 28/37 (75%), Gaps = 3/37 (8%)
Frame = +2
Query: 300 SPFSKSNANFTKMVV--SPLDELC-VESIHHKASIEV 333
SPFS ++A+FTKMV SPLDEL +S+ HKA I+V
Sbjct: 2 SPFSSTDADFTKMVEVNSPLDELLYADSVFHKAFIKV 112
>TC15011 similar to UP|Q8GT65 (Q8GT65) Serpin-like protein (Fragment),
partial (20%)
Length = 623
Score = 88.2 bits (217), Expect = 2e-18
Identities = 48/86 (55%), Positives = 60/86 (68%), Gaps = 2/86 (2%)
Frame = +2
Query: 305 SNANFTKMVVSPL--DELCVESIHHKASIEVNEEGTEAGVACSMCDVKLCVSARTGIDFV 362
++A+ TKMV S L V +I HK+ IEVNEEGTEA A ++ + T +DFV
Sbjct: 5 NDAHLTKMVDSSTVGQGLYVSNIFHKSFIEVNEEGTEAAAASAVTIKLRSLQVLTRVDFV 184
Query: 363 ADHPFLFLIREDLTGTILFIGQVLHP 388
ADHPFLF+IREDLTGT+LF GQVL+P
Sbjct: 185 ADHPFLFVIREDLTGTVLFTGQVLNP 262
>BP048545
Length = 452
Score = 81.6 bits (200), Expect = 2e-16
Identities = 46/79 (58%), Positives = 57/79 (71%), Gaps = 1/79 (1%)
Frame = -2
Query: 311 KMVVSPLDELCVESIHHKASIEVNEEGTEAGVACSMCDVKLCVSAR-TGIDFVADHPFLF 369
KMV S +L V +I HK+ IEVNEEGT A A + +K +S R T +DFVADHPF F
Sbjct: 451 KMVDS---DLYVSNIFHKSFIEVNEEGTRAAAASAGIVLKRSLSPRPTRVDFVADHPFFF 281
Query: 370 LIREDLTGTILFIGQVLHP 388
+IRE++TGTILF GQVL+P
Sbjct: 280 VIREEMTGTILFTGQVLNP 224
>AU088851
Length = 663
Score = 78.2 bits (191), Expect = 2e-15
Identities = 51/99 (51%), Positives = 60/99 (60%), Gaps = 3/99 (3%)
Frame = +3
Query: 49 GSAGSTLDELFSFLRFDSVDHLNTFFSQVLSTVL---SDTTPSFLLSFVNEMWADKSLAL 105
GS GSTLDEL SFLRFDS D LN+F SQ++S VL PS LSF + MWADK+L L
Sbjct: 267 GSEGSTLDELLSFLRFDSTDRLNSFTSQLVSDVLYVNDAAPPSPRLSFASRMWADKTLFL 446
Query: 106 SHSFKQLMTTHYKATLALVDFRTKGDQVCREVNSWVEKE 144
S L K LA +DF +G + CR V WV K+
Sbjct: 447 SLPSYNLWLL-IKELLASLDFXKQGKKXCR-V*IWVGKK 557
Score = 68.9 bits (167), Expect = 1e-12
Identities = 40/88 (45%), Positives = 52/88 (58%)
Frame = +1
Query: 1 MDLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFS 60
MDL++S Q++VAL++TKHLFSKE YQ +NLVFSP SLH +LS+MA
Sbjct: 121 MDLKESTSHQSDVALTLTKHLFSKEVYQHENLVFSPLSLHVVLSIMAP------------ 264
Query: 61 FLRFDSVDHLNTFFSQVLSTVLSDTTPS 88
HL +FF ST + +TPS
Sbjct: 265 --LVQKAPHLMSFFPSFDSTPPTVSTPS 342
>BP034134
Length = 556
Score = 69.3 bits (168), Expect = 1e-12
Identities = 35/41 (85%), Positives = 38/41 (92%), Gaps = 1/41 (2%)
Frame = -1
Query: 357 TGIDFVADHPFLFLIREDLTGTILFIGQVLHP-EGAAMSLK 396
TGIDF ADHPFLFLIREDLTGTILFIGQVL+P +GAA S+K
Sbjct: 487 TGIDFEADHPFLFLIREDLTGTILFIGQVLNPLDGAAASVK 365
>TC19574 weakly similar to UP|Q8GT65 (Q8GT65) Serpin-like protein
(Fragment), partial (8%)
Length = 414
Score = 66.2 bits (160), Expect = 9e-12
Identities = 34/46 (73%), Positives = 39/46 (83%), Gaps = 1/46 (2%)
Frame = -3
Query: 358 GIDFVADHPFLFLIREDLTGTILFIGQVLHP-EGAAMSLKVSNFYI 402
GIDFVA+HPFLFLIRE+LTGT LFIGQVL+P +GA +KV NF I
Sbjct: 355 GIDFVANHPFLFLIREELTGTFLFIGQVLNPLDGADPLVKVCNFNI 218
>BP073359
Length = 388
Score = 65.1 bits (157), Expect = 2e-11
Identities = 34/53 (64%), Positives = 40/53 (75%), Gaps = 2/53 (3%)
Frame = -3
Query: 339 EAGVACSMCDVKLCVS--ARTGIDFVADHPFLFLIREDLTGTILFIGQVLHPE 389
EA A S V++C S T DFVADHPFLFLI+EDL+GTILFIG+VLHP+
Sbjct: 386 EAAAASSGRLVRMCDSDSVSTDTDFVADHPFLFLIKEDLSGTILFIGRVLHPD 228
>TC10025 weakly similar to UP|Q8GT65 (Q8GT65) Serpin-like protein
(Fragment), partial (15%)
Length = 672
Score = 64.7 bits (156), Expect = 3e-11
Identities = 37/66 (56%), Positives = 44/66 (66%), Gaps = 1/66 (1%)
Frame = +1
Query: 332 EVNEEGTEAGVACSMCDVKLCVSARTGIDFVADHPFLFLIREDLTGTILFIGQVLHP-EG 390
EVNEE T A A + V IDFVADHPFL L+RED+TGTILF+GQVL+P +G
Sbjct: 1 EVNEEETVAVAATAARMVLGGAGLCPHIDFVADHPFLLLVREDITGTILFVGQVLNPLDG 180
Query: 391 AAMSLK 396
A +K
Sbjct: 181 AGTPVK 198
>BP051691
Length = 428
Score = 49.3 bits (116), Expect(2) = 5e-11
Identities = 27/48 (56%), Positives = 33/48 (68%), Gaps = 5/48 (10%)
Frame = -2
Query: 303 SKSNANFTKMVVSPLDE-----LCVESIHHKASIEVNEEGTEAGVACS 345
S+ +ANFT MV SP DE L ++++ HKA IEVNEEGTEA A S
Sbjct: 427 SRCDANFTNMVDSPSDEFPSDELYIDNMFHKAFIEVNEEGTEAAAASS 284
Score = 34.3 bits (77), Expect(2) = 5e-11
Identities = 14/17 (82%), Positives = 17/17 (99%)
Frame = -1
Query: 373 EDLTGTILFIGQVLHPE 389
EDL+GTILFIG+VLHP+
Sbjct: 275 EDLSGTILFIGRVLHPD 225
>TC12056 weakly similar to UP|ILEU_HUMAN (P30740) Leukocyte elastase
inhibitor (LEI) (Monocyte/neutrophil elastase inhibitor)
(M/NEI) (EI), partial (6%)
Length = 401
Score = 62.4 bits (150), Expect = 1e-10
Identities = 28/31 (90%), Positives = 29/31 (93%)
Frame = +2
Query: 360 DFVADHPFLFLIREDLTGTILFIGQVLHPEG 390
+FVADHPFLFLIRED TGTILFIGQVLHP G
Sbjct: 2 NFVADHPFLFLIREDFTGTILFIGQVLHPHG 94
>TC9744 similar to UP|Q40066 (Q40066) Protein zx, partial (7%)
Length = 527
Score = 61.6 bits (148), Expect = 2e-10
Identities = 36/63 (57%), Positives = 43/63 (68%), Gaps = 1/63 (1%)
Frame = +1
Query: 336 EGTEAGVACSMCDVKLCVSARTGIDFVADHPFLFLIREDLTGTILFIGQVLHP-EGAAMS 394
+GTEA A + K + G FVADHPFLFLIRED TGTILF+GQVL+P EGA +
Sbjct: 1 KGTEATSATVVSGRKR-LGGFLGTYFVADHPFLFLIREDFTGTILFVGQVLNPLEGAPVK 177
Query: 395 LKV 397
K+
Sbjct: 178 KKI 186
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.323 0.136 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,653,382
Number of Sequences: 28460
Number of extensions: 87635
Number of successful extensions: 429
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 413
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 413
length of query: 402
length of database: 4,897,600
effective HSP length: 92
effective length of query: 310
effective length of database: 2,279,280
effective search space: 706576800
effective search space used: 706576800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0175.7


