

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0175.13
(437 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
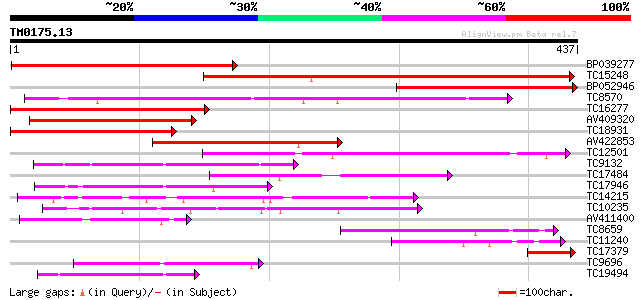
Score E
Sequences producing significant alignments: (bits) Value
BP039277 353 3e-98
TC15248 weakly similar to UP|Q8GT21 (Q8GT21) Benzoyl coenzyme A:... 313 3e-86
BP052946 286 4e-78
TC8570 similar to UP|Q9FLM5 (Q9FLM5) N-hydroxycinnamoyl/benzoylt... 244 3e-65
TC16277 weakly similar to UP|Q43583 (Q43583) Hsr201 protein, par... 200 4e-52
AV409320 198 1e-51
TC18931 similar to UP|Q8GT20 (Q8GT20) Benzoyl coenzyme A: benzyl... 182 7e-47
AV422853 174 3e-44
TC12501 similar to UP|Q9LR83 (Q9LR83) F21B7.2, partial (55%) 130 4e-31
TC9132 similar to UP|Q8GSM7 (Q8GSM7) Hydroxycinnamoyl transferas... 112 1e-25
TC17484 weakly similar to UP|DBAT_TAXCU (Q9M6E2) 10-deacetylbacc... 101 3e-22
TC17946 similar to UP|Q9FI59 (Q9FI59) Gb|AAD29063.1, partial (6%) 91 4e-19
TC14215 similar to UP|Q9FYM1 (Q9FYM1) F21J9.8, partial (8%) 84 4e-17
TC10235 79 1e-15
AV411400 75 2e-14
TC8659 similar to UP|Q8VWP8 (Q8VWP8) Acyltransferase-like protei... 62 1e-10
TC11240 similar to UP|Q8GSM7 (Q8GSM7) Hydroxycinnamoyl transfera... 59 2e-09
TC17379 homologue to UP|Q8G4D2 (Q8G4D2) Probable ribosomal-prote... 50 7e-07
TC9696 weakly similar to UP|Q8LAF8 (Q8LAF8) Fatty acid elongase-... 50 7e-07
TC19494 weakly similar to UP|Q9SND9 (Q9SND9) Anthranilate N-hydr... 45 2e-05
>BP039277
Length = 522
Score = 353 bits (906), Expect = 3e-98
Identities = 173/174 (99%), Positives = 173/174 (99%)
Frame = +1
Query: 2 DSSPPSLTFKVQRCKPELVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADK 61
DSSPPSLTFKVQRCKPELVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADK
Sbjct: 1 DSSPPSLTFKVQRCKPELVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADK 180
Query: 62 DPVQAIRQALSRTLVYYYPFAGRLKEGPGRKLMVDCTGEGVMFIEANADVSLVEFGETPL 121
DPVQAIRQALSRTLVYYYPFAGRLKEG GRKLMVDCTGEGVMFIEANADVSLVEFGETPL
Sbjct: 181 DPVQAIRQALSRTLVYYYPFAGRLKEGLGRKLMVDCTGEGVMFIEANADVSLVEFGETPL 360
Query: 122 PPFPCFEELLYDVPGSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLK 175
PPFPCFEELLYDVPGSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLK
Sbjct: 361 PPFPCFEELLYDVPGSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLK 522
>TC15248 weakly similar to UP|Q8GT21 (Q8GT21) Benzoyl coenzyme A: benzyl
alcohol benzoyl transferase, partial (52%)
Length = 1085
Score = 313 bits (803), Expect = 3e-86
Identities = 150/293 (51%), Positives = 205/293 (69%), Gaps = 7/293 (2%)
Frame = +1
Query: 150 VTRLKCGGFIMALRFNHTMSDGAGLKQFLSALAEMAQGASQPLTPPVWCRELLMARDPPR 209
VTRLKCGGFI ALRFNH M+DGAG+ F+ A+ E+A+GA++P PVW RELL ARDPPR
Sbjct: 13 VTRLKCGGFIFALRFNHVMTDGAGIVHFMYAVVEIARGANEPSILPVWQRELLHARDPPR 192
Query: 210 ITCNHYEFEQVPSDSTEEGAIT------RSFFFGSNEIAALRRLVPLDL-RHCSTFDVIT 262
+T NH E+EQ+ D+ + +T +SFFFG EIAA+RR +P L +T++V+T
Sbjct: 193 VTHNHREYEQLTDDTITDPILTGTDFVQQSFFFGPAEIAAIRRHLPHHLDTESTTYEVLT 372
Query: 263 ACFWYCRTKALQLAPHDDIRLMTIVNTRNRFNPPIPVGYYGNCFTYPAVVTTVGKLCGNS 322
+ W CRTKALQL P ++R+M I + R +FNPP P GYYGNCF +PA V T G+LC
Sbjct: 373 SYIWRCRTKALQLDPSQEVRMMCITDARGKFNPPFPTGYYGNCFAFPAAVATAGELCEKP 552
Query: 323 FGYAVELVRKAKAQATEEYMHSMADFLVANRRCLFTTVRSCIVSDLTRFKLHETDFGWGE 382
+AV L++KA + +EEYMHS+AD +V + LFT VRSC+V D T E DFGWG+
Sbjct: 553 LEHAVRLIKKASGEMSEEYMHSLADLMVTEGKPLFTVVRSCVVLDTTYAGFRELDFGWGK 732
Query: 383 PVCGGVAKGGAGLYGGASYIIACKNAKGEDGRVLVICLPVENMKRFAKELNNM 435
V GG+A+ GAG + ++ + +NA+GE+G ++++CLP + M FAKEL++M
Sbjct: 733 AVYGGLAQAGAGAFPAVNFHVPSQNAQGEEGILVLVCLPAKIMSVFAKELDDM 891
>BP052946
Length = 559
Score = 286 bits (733), Expect = 4e-78
Identities = 137/139 (98%), Positives = 138/139 (98%)
Frame = -1
Query: 299 VGYYGNCFTYPAVVTTVGKLCGNSFGYAVELVRKAKAQATEEYMHSMADFLVANRRCLFT 358
VGYYGNCFTYPAVVTTVGKLCGNSFGYAVELVRKAKAQATEEYMHSM DFLVANRRCLFT
Sbjct: 559 VGYYGNCFTYPAVVTTVGKLCGNSFGYAVELVRKAKAQATEEYMHSMXDFLVANRRCLFT 380
Query: 359 TVRSCIVSDLTRFKLHETDFGWGEPVCGGVAKGGAGLYGGASYIIACKNAKGEDGRVLVI 418
TVRSCIVSDLTRFKLHETDFGWGEPVCGGVA+GGAGLYGGASYIIACKNAKGEDGRVLVI
Sbjct: 379 TVRSCIVSDLTRFKLHETDFGWGEPVCGGVAQGGAGLYGGASYIIACKNAKGEDGRVLVI 200
Query: 419 CLPVENMKRFAKELNNMIV 437
CLPVENMKRFAKELNNMIV
Sbjct: 199 CLPVENMKRFAKELNNMIV 143
>TC8570 similar to UP|Q9FLM5 (Q9FLM5)
N-hydroxycinnamoyl/benzoyltransferase-like protein,
partial (91%)
Length = 1551
Score = 244 bits (622), Expect = 3e-65
Identities = 143/387 (36%), Positives = 211/387 (53%), Gaps = 11/387 (2%)
Frame = +1
Query: 12 VQRCKPELVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADKDPVQA---IR 68
V+ +P LV PA T + LS++D NI I+ + A+K +A ++
Sbjct: 94 VKLSEPALVLPAEETKKGMYFLSNLDQ------NIAVIIRTVYCFKTAEKGNEKAGEVVK 255
Query: 69 QALSRTLVYYYPFAGRLKEGPGRKLMVDCTGEGVMFIEANADVSLVEFGETPLPPFPCFE 128
AL + LV+YYP AGRL P KL+V+CTGEG +F+EA A+ SL E G+ P
Sbjct: 256 SALKKVLVHYYPLAGRLSISPEGKLIVECTGEGALFVEAEANCSLEEIGDITKPDPGTLG 435
Query: 129 ELLYDVPGSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFLSALAEMAQGA 188
+L+YD+PG++ +L P L+ QVT+ KCGGF + L NH M DG G +F+++ E A+
Sbjct: 436 KLVYDIPGAKHILQMPPLVAQVTKFKCGGFSLGLCMNHCMFDGIGAMEFVNSWGEAAR-- 609
Query: 189 SQPLT-PPVWCRELLMARDPPRITCNHYEFEQVPSDST-----EEGAITRSFFFGSNEIA 242
+ PL+ PP+ R +L AR+PP+I H EF + S E+ + RSF F ++
Sbjct: 610 ALPLSIPPILDRSILKARNPPKIEHLHQEFADIEDKSNTNTLYEDEMLYRSFCFDPEKLK 789
Query: 243 ALRRLVPLD--LRHCSTFDVITACFWYCRTKALQLAPHDDIRLMTIVNTRNRFNPPIPVG 300
L+ D L C+TF+V++A W RTKAL+L P + +L+ V+ R F PP+P G
Sbjct: 790 QLKMKAMEDGALESCTTFEVLSAFVWIARTKALKLLPEQETKLLFAVDGRKNFTPPLPKG 969
Query: 301 YYGNCFTYPAVVTTVGKLCGNSFGYAVELVRKAKAQATEEYMHSMADFLVANRRCLFTTV 360
Y+GN V G+L +AV L++ A T+ YM S D+ R +
Sbjct: 970 YFGNGIVLTNSVCQAGELSEKKLSHAVRLIQDAVKMVTDSYMRSAIDYFEVT-RARPSLA 1146
Query: 361 RSCIVSDLTRFKLHETDFGWGEPVCGG 387
+ +++ +R H TDFGWGEPV G
Sbjct: 1147CTLLITTWSRLGFHTTDFGWGEPVLSG 1227
>TC16277 weakly similar to UP|Q43583 (Q43583) Hsr201 protein, partial (32%)
Length = 547
Score = 200 bits (508), Expect = 4e-52
Identities = 97/155 (62%), Positives = 121/155 (77%), Gaps = 1/155 (0%)
Frame = +1
Query: 1 MDSSPPSLTFKVQRCKPELVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMAD 60
++ S P L F V+R +PELV PA STPHE K LSDIDDQ GLR N+P I YR+EPSMA
Sbjct: 82 LEPSSPPLVFSVRRREPELVAPAVSTPHETKHLSDIDDQAGLRANVPIIQFYRNEPSMAG 261
Query: 61 KDPVQAIRQALSRTLVYYYPFAGRLKEGPGRKLMVDCTGEGVMFIEANADVSLVEFGETP 120
KDPV IR A+++ LV+YYP AGRL+E G L+VDC EGVMFIEA+AD++ +FG+T
Sbjct: 262 KDPVDIIRNAVAKALVFYYPLAGRLREAAGGNLVVDCNEEGVMFIEADADITFEQFGDTL 441
Query: 121 LPPFPCFEELLYDVPGSE-QVLDSPLLLIQVTRLK 154
PPFPCF+ELL++ PGSE V++SP++LIQVTRLK
Sbjct: 442 KPPFPCFQELLHEAPGSEGVVINSPMILIQVTRLK 546
>AV409320
Length = 392
Score = 198 bits (504), Expect = 1e-51
Identities = 93/129 (72%), Positives = 111/129 (85%)
Frame = +3
Query: 16 KPELVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADKDPVQAIRQALSRTL 75
+PELV PA +TP EVKLLSDIDDQDGLRF IP YR+ PSMA KDPV AIR+AL++TL
Sbjct: 6 EPELVGPAEATPREVKLLSDIDDQDGLRFQIPVAQFYRYNPSMAGKDPVDAIRKALAKTL 185
Query: 76 VYYYPFAGRLKEGPGRKLMVDCTGEGVMFIEANADVSLVEFGETPLPPFPCFEELLYDVP 135
V+YYPFAGRL+EGPGRKLMVDCT EGV+FIEA+ADV+L EFG+ PFPC EELLY+VP
Sbjct: 186 VFYYPFAGRLREGPGRKLMVDCTAEGVLFIEADADVTLNEFGDNLQTPFPCMEELLYEVP 365
Query: 136 GSEQVLDSP 144
GS+++L++P
Sbjct: 366 GSDEMLNTP 392
>TC18931 similar to UP|Q8GT20 (Q8GT20) Benzoyl coenzyme A: benzyl alcohol
benzoyl transferase, partial (26%)
Length = 446
Score = 182 bits (463), Expect = 7e-47
Identities = 84/128 (65%), Positives = 105/128 (81%)
Frame = +3
Query: 1 MDSSPPSLTFKVQRCKPELVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMAD 60
M SSP SL F V+RC+PELV P+ TP E+KLLSDIDDQ+GLRF IP I YR++PSM
Sbjct: 63 MASSPASLAFTVRRCEPELVAPSQPTPRELKLLSDIDDQEGLRFQIPVIQFYRYDPSMTG 242
Query: 61 KDPVQAIRQALSRTLVYYYPFAGRLKEGPGRKLMVDCTGEGVMFIEANADVSLVEFGETP 120
KDPV+ IR+AL++TLV+YYP AGRL EGP RKLMV+CT +GV+FIEA+ADV++ +FG+
Sbjct: 243 KDPVEVIRKALAKTLVFYYPLAGRLMEGPDRKLMVNCTADGVLFIEADADVTMKQFGDAL 422
Query: 121 LPPFPCFE 128
PPFPC+E
Sbjct: 423 QPPFPCWE 446
>AV422853
Length = 456
Score = 174 bits (440), Expect = 3e-44
Identities = 84/151 (55%), Positives = 108/151 (70%), Gaps = 5/151 (3%)
Frame = +1
Query: 111 VSLVEFGETPLPPFPCFEELLYDVPGSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMSD 170
++L +FG+ PFPC +ELLY+VPGSE++L++PLLLIQVTRLKCGGFI A+R NHTM D
Sbjct: 1 ITLNQFGDNLQTPFPCMDELLYEVPGSEEMLNTPLLLIQVTRLKCGGFIFAIRLNHTMCD 180
Query: 171 GAGLKQFLSALAEMAQGASQPLTPPVWCRELLMARDPPRITCNHYEF-EQVP----SDST 225
GL QFLSA+ EMA+G QP PVWCRE+L ARD PR+TC H E+ EQVP +
Sbjct: 181 AVGLVQFLSAIGEMARGMPQPSILPVWCREILSARDSPRVTCTHPEYDEQVPYPKETTIP 360
Query: 226 EEGAITRSFFFGSNEIAALRRLVPLDLRHCS 256
++ + SFFFG NE+A +R +P C+
Sbjct: 361 QDDMVHESFFFGPNELATIRSFLPSHQLRCT 453
>TC12501 similar to UP|Q9LR83 (Q9LR83) F21B7.2, partial (55%)
Length = 1062
Score = 130 bits (327), Expect = 4e-31
Identities = 90/303 (29%), Positives = 138/303 (44%), Gaps = 19/303 (6%)
Frame = +3
Query: 149 QVTRLKCGGFIMALRFNHTMSDGAGLKQFLSALAEMAQGASQPLTP-PVWCRELLMARDP 207
QVT+ +CGGF + LR H + DG G QFL+A A A+ S + P P W RE+ R P
Sbjct: 48 QVTQFRCGGFSLGLRLCHCICDGLGAMQFLAAWAATAKSGSLVIDPKPCWDREMFKPRHP 227
Query: 208 PRITCNHYEFEQVPSDSTEEGAITRSFFFGSNEIAALRRL------------VPLDLRHC 255
P + H EF ++ EEG+ + + + R+ P D C
Sbjct: 228 PMVKFPHMEFMRI-----EEGSNLTITLWQTKPVQKCYRIQREFQNYLKTLAQPSDAAGC 392
Query: 256 STFDVITACFWYCRTKALQLAPHD-DIRLMTIVNTRNRF-NPPIPVGYYGNCFTYPAVVT 313
+TFD + A W KAL + P D +RL VN R + NPP+ G+YGN +
Sbjct: 393 TTFDAMAAHIWRSWVKALDVRPLDYTLRLTFSVNARPKLRNPPLREGFYGNVVCVACTTS 572
Query: 314 TVGKLCGNSFGYAVELVRKAKAQATEEYMHSMADFLVANRRCLFTTVRSCIVSDLTRFKL 373
+V +L LVR+A+ +EEY+ S D++ +R ++ TRF +
Sbjct: 573 SVSELVHGQLPETTRLVREARQSVSEEYLRSTVDYVDVDRPKQLEFGGKLTITQWTRFSM 752
Query: 374 HET-DFGWGEPVCGGVAKGGAGLYGGASYIIACKNAKGED---GRVLVICLPVENMKRFA 429
+++ DFGWG+P+ G L + + D ++ ICLP ++F
Sbjct: 753 YKSADFGWGKPLYA----GPIDLTPTPQVCVFLPEGEAADCSGSMIVCICLPESAAQKFT 920
Query: 430 KEL 432
+ L
Sbjct: 921 QAL 929
>TC9132 similar to UP|Q8GSM7 (Q8GSM7) Hydroxycinnamoyl transferase, partial
(53%)
Length = 878
Score = 112 bits (280), Expect = 1e-25
Identities = 68/204 (33%), Positives = 109/204 (53%)
Frame = +1
Query: 19 LVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADKDPVQAIRQALSRTLVYY 78
+V PA P S++D F+ P + YR + D + +++ALS+ LV +
Sbjct: 175 MVRPAEEVPQRTVWNSNVDLVVP-NFHTPSVYFYRANGASNFFD-AKVLKEALSKVLVPF 348
Query: 79 YPFAGRLKEGPGRKLMVDCTGEGVMFIEANADVSLVEFGETPLPPFPCFEELLYDVPGSE 138
YP AGRL+ ++ +DC G+GV+F+EA+ + +FG+ P +L+ V S
Sbjct: 349 YPMAGRLRRDDDGRVEIDCDGQGVLFVEADTGAVIDDFGD--FAPTLQQRQLIPGVDYSR 522
Query: 139 QVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFLSALAEMAQGASQPLTPPVWC 198
+ PLL++QVT KCGG + + H ++DGA F++ +++A+G + PP
Sbjct: 523 GIESYPLLVLQVTHFKCGGVSLGVGMQHHVADGASGLHFINTWSDVARGLDVSI-PPFID 699
Query: 199 RELLMARDPPRITCNHYEFEQVPS 222
R LL ARDPPR H E++ PS
Sbjct: 700 RTLLRARDPPRPAFEHIEYKPPPS 771
>TC17484 weakly similar to UP|DBAT_TAXCU (Q9M6E2) 10-deacetylbaccatin III
10-O-acetyltransferase (DBAT) , partial (11%)
Length = 577
Score = 101 bits (251), Expect = 3e-22
Identities = 55/200 (27%), Positives = 96/200 (47%), Gaps = 13/200 (6%)
Frame = +3
Query: 155 CGGFIMALRFNHTMSDGAGLKQFLSALAEMAQGASQPLTPPVWCRELLMARD-------- 206
CGGF++ L F H++ DG G QFL+A+ E+A+G ++P P W R+ ++
Sbjct: 9 CGGFVIGLMFCHSICDGLGAAQFLNAVGELARGLNKPTIEPAWHRDFFPPQEAEAEAQPN 188
Query: 207 -----PPRITCNHYEFEQVPSDSTEEGAITRSFFFGSNEIAALRRLVPLDLRHCSTFDVI 261
PP + + + + + SF + + CS F+++
Sbjct: 189 LKLLGPPPMPDYQLQHANIDMPMDQIKQLKSSFEQVTGQT-------------CSAFEIV 329
Query: 262 TACFWYCRTKALQLAPHDDIRLMTIVNTRNRFNPPIPVGYYGNCFTYPAVVTTVGKLCGN 321
A FW RT+A+ L P+ ++L+ N RN NPP+P G+YGNCF + + L
Sbjct: 330 AAKFWSSRTRAIDLDPNTQLKLVFFANCRNLLNPPLPKGFYGNCFFPVTITASCEFLRDA 509
Query: 322 SFGYAVELVRKAKAQATEEY 341
+ V+++++AK + E+
Sbjct: 510 TIIEVVKVIQEAKGELPLEF 569
>TC17946 similar to UP|Q9FI59 (Q9FI59) Gb|AAD29063.1, partial (6%)
Length = 699
Score = 90.9 bits (224), Expect = 4e-19
Identities = 57/186 (30%), Positives = 102/186 (54%), Gaps = 3/186 (1%)
Frame = +3
Query: 20 VPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADKDPVQAIRQALSRTLVYYY 79
VPP TPH LSD+D R + P + +Y+ + D+ ++ ++ +LS+ LV+YY
Sbjct: 144 VPPNQPTPH*HLWLSDMDQVSRQR-HTPIVYIYK---AKHDQYCIERMKDSLSKILVHYY 311
Query: 80 PFAGRLKEGPGRKLMVDCTGEGVMFIEANADVSLVEFGETPLPPFPCFEELLYDVPGSEQ 139
P AGR ++ ++C +GV+ +EA + ++ +FG+ P +EL+ + +
Sbjct: 312 PVAGRFSFTENGRMEINCNAKGVILLEAETEKTMADFGD--FSPSDSTKELVPTIDYNHP 485
Query: 140 VLDSPLLLIQVTRLKC--GGFIMALRFNHTMSDGAGLKQFLSALAEMAQGAS-QPLTPPV 196
+ + PLL +Q+TR GG + + HT+SDG G +F ++ A++A+G + +P P
Sbjct: 486 IEELPLLAVQLTRFNGGEGGLADGVAWVHTLSDGLGCTRFFNSWAKIARGDTLEPHEKPS 665
Query: 197 WCRELL 202
R LL
Sbjct: 666 LDRTLL 683
>TC14215 similar to UP|Q9FYM1 (Q9FYM1) F21J9.8, partial (8%)
Length = 1096
Score = 84.3 bits (207), Expect = 4e-17
Identities = 87/324 (26%), Positives = 142/324 (42%), Gaps = 15/324 (4%)
Frame = +1
Query: 7 SLTFKVQRCKPELVPPASSTPHEVKL--LSDIDDQDGLRFNIPFIMMYRHEPSMADKDPV 64
S T +V+ E++ P++ TP +++ LS ID+ R IP + Y P+ +DK +
Sbjct: 85 SATMEVELISTEIIKPSTPTPPHLRIYPLSFIDNMM-YRNYIPVALFYH--PNESDKQSI 255
Query: 65 QA-IRQALSRTLVYYYPFAGRLKEGPGRKLMVDCTGEGVMF----IEANADVSLVEFGET 119
+ ++ +LS L YYPFAGRL++ +L ++C EGV F I+ L ET
Sbjct: 256 ISNLKNSLSEVLTRYYPFAGRLRD----QLSIECNDEGVPFRVTEIKGEQSTILQNPNET 423
Query: 120 PLPPFPCFEELLY--DVPGSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQF 177
L LL+ +P D + +Q+ CGG + H M D F
Sbjct: 424 LL-------RLLFPDKLPWKVTEYDESIAAVQINFFSCGGLAITACVCHKMGDATTTLNF 582
Query: 178 LSALAEMAQGASQPLTP---PVWCR--ELLMARDPPRITCNHYEFEQVPSDSTEEGAITR 232
++ A M + + LT P+ + +D P E V + A+ R
Sbjct: 583 VNDWAAMTKEKGEALTQLSLPLLNGGVSVFPHKDMPCFP------EIVFGKGNKSNAVCR 744
Query: 233 SFFFGSNEIAALRRLVPL-DLRHCSTFDVITACFWYCRTKALQLAPHDDIRLMTIVNTRN 291
F F ++I +L+ +V +++ + +VITA + AL L D +V+ R
Sbjct: 745 RFVFPPSKINSLKEMVTSHGVQNPTRVEVITAWIYMRAVHALGLT-FDKTSFRQVVSLRR 921
Query: 292 RFNPPIPVGYYGNCFTYPAVVTTV 315
R PP+P GN F + + T V
Sbjct: 922 RMTPPLPNKSVGNMFWFLYMCTPV 993
>TC10235
Length = 1116
Score = 79.0 bits (193), Expect = 1e-15
Identities = 80/317 (25%), Positives = 131/317 (41%), Gaps = 24/317 (7%)
Frame = +1
Query: 26 TPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADKDPVQAIRQALSRTLVYYYPFAGRL 85
TP ++ LL+ Q GL F H+P + + ++ LS L ++ PF GRL
Sbjct: 163 TPWDLLLLNLESIQQGLLF---------HKPKTDHQ--IHHLKHTLSTILTFFPPFTGRL 309
Query: 86 --------KEGPGRKLMVDCTGEGVMFIEANADVSLVEFGETPLPPF-PCFEELLYDVPG 136
+ C G +F+ A A+ + + + P + P + + G
Sbjct: 310 IITNHHDNNNNTTVTCHITCNNAGALFVHAAAEKTSI--ADIIRPGYVPSIVHSFFPLNG 483
Query: 137 SE--QVLDSPLLLIQVTRLKCGGFIMALRFNHTMSDGAGLKQFLSALAEMAQGASQPL-- 192
+ Q + PLL +QVT L G FI NH+++DG QF+++ E+++G S L
Sbjct: 484 VKNYQGMSQPLLAVQVTELLDGVFI-GFTMNHSVADGKAFWQFINSWGELSRGNSDKLMK 660
Query: 193 TPPVWCRELLMARDP----PRITCNHYEFEQVPSDSTEEGAITRSFFFGSNEIAALRRLV 248
TPP+ R DP P T + +Q SD + F F ++A L+
Sbjct: 661 TPPLE-RWFPNGIDPPIHFPSFTKEEEKKKQDNSDPVPQLPPLCIFHFSKEKLAELKAKA 837
Query: 249 PLDL-------RHCSTFDVITACFWYCRTKALQLAPHDDIRLMTIVNTRNRFNPPIPVGY 301
++ R S+ + W +A ++ P + M I++ R R PP+P Y
Sbjct: 838 NAEVDNGMHENRVISSLQALLTHIWRSVIRAQRVNPEKETDYMLIIDARQRMQPPLPENY 1017
Query: 302 YGNCFTYPAVVTTVGKL 318
GN V G+L
Sbjct: 1018LGNAGMAGIVKLKAGEL 1068
>AV411400
Length = 417
Score = 75.1 bits (183), Expect = 2e-14
Identities = 52/137 (37%), Positives = 74/137 (53%), Gaps = 4/137 (2%)
Frame = +2
Query: 8 LTFKVQRCKPELVPPASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADKDPVQAI 67
+ V R K +V PA TP LS +D LR N + ++RH P + + I
Sbjct: 23 MAMSVIRTKRGMVKPAKETPLTTLDLSVMDRLPVLRCNARTLHVFRHGP-----EATRVI 187
Query: 68 RQALSRTLVYYYPFAGRLKEGPGRKLMVDCTGEGVMFIEANADVSLVE---FGETPLPPF 124
R+ALS LV YYP AGRL E R L ++C+GEGV ++EA+AD +L F + P
Sbjct: 188 REALSLALVPYYPLAGRLIESKPRCLQIECSGEGVWYVEASADCTLHSVNFFDDVQSIP- 364
Query: 125 PCFEELLYD-VPGSEQV 140
+++LL D +P +E V
Sbjct: 365 --YDDLLPDHIPENEHV 409
>TC8659 similar to UP|Q8VWP8 (Q8VWP8) Acyltransferase-like protein
(Fragment), partial (42%)
Length = 866
Score = 62.4 bits (150), Expect = 1e-10
Identities = 48/171 (28%), Positives = 74/171 (43%), Gaps = 3/171 (1%)
Frame = +3
Query: 256 STFDVITACFWYCRTKALQLAPHDDIRLMTIVNTRNRFNPPIPVGYYGNCFTYPAVVTTV 315
STF ++ W T A L P D V+ R R +PP+P Y+GN VT V
Sbjct: 3 STFQALSTHIWRHVTHARSLKPEDYTVFTVFVDCRKRVDPPMPEAYFGNLIQAIFTVTAV 182
Query: 316 GKLCGNSFGYAVELVRKA-KAQATEEYMHSMADFLVANRRCLF--TTVRSCIVSDLTRFK 372
G L + + +++KA +A + D+ A + F + V RFK
Sbjct: 183 GLLSAHPPQFGATMIQKAIEAHDAKAIDERNKDWESAPKIFQFKDAGINCVTVGSSPRFK 362
Query: 373 LHETDFGWGEPVCGGVAKGGAGLYGGASYIIACKNAKGEDGRVLVICLPVE 423
+++ DFGWG+P V G + G Y+ K+ GR + + L +E
Sbjct: 363 VYDIDFGWGKPEI--VRSGPNNKFDGMIYLYPGKSG----GRSIDVELTLE 497
>TC11240 similar to UP|Q8GSM7 (Q8GSM7) Hydroxycinnamoyl transferase, partial
(31%)
Length = 528
Score = 58.9 bits (141), Expect = 2e-09
Identities = 45/142 (31%), Positives = 60/142 (41%), Gaps = 8/142 (5%)
Frame = +1
Query: 295 PPIPVGYYGNCFTYPAVVTTVGKLCGNSFGYAVELVRKAKAQATEEYMHSMADF--LVAN 352
PP+P GY+GN + G L YA + A + EY+ S DF L +
Sbjct: 28 PPLPQGYFGNVIFTATPIAMAGDLMSKPIWYAASRIHDALLRMDNEYLRSALDFLELQPD 207
Query: 353 RRCLFTTVRSCIVSDL-----TRFKLHETDFGWGEPVCGGVAKGGAGLYGGASYIIACKN 407
R L + +L R +H+ DFGWG P+ G G Y G S+II
Sbjct: 208 LRALVRGAHTFRCPNLGITSWVRLPIHDADFGWGRPIFMG---PGGIAYEGLSFIIP--- 369
Query: 408 AKGEDGRVLV-ICLPVENMKRF 428
+ DG + V I L E MK F
Sbjct: 370 SPVNDGSLSVAIALQPEQMKVF 435
>TC17379 homologue to UP|Q8G4D2 (Q8G4D2) Probable ribosomal-protein-alanine
N-acetyltransferase, partial (6%)
Length = 519
Score = 50.1 bits (118), Expect = 7e-07
Identities = 22/37 (59%), Positives = 30/37 (80%)
Frame = +3
Query: 400 SYIIACKNAKGEDGRVLVICLPVENMKRFAKELNNMI 436
S+ IA KNAKGE+G V+ +CLP E M+RF KEL+N++
Sbjct: 3 SFYIAFKNAKGEEGLVIPVCLPSEAMERFTKELDNVL 113
>TC9696 weakly similar to UP|Q8LAF8 (Q8LAF8) Fatty acid elongase-like
protein (cer2-like), partial (23%)
Length = 881
Score = 50.1 bits (118), Expect = 7e-07
Identities = 33/148 (22%), Positives = 64/148 (42%), Gaps = 2/148 (1%)
Frame = +3
Query: 50 MMYRHEPSMADKDPVQAIRQALSRTLVYYYPFAGRLKEGPGRKLMVDCTGEGVMFIEANA 109
++Y + A I++ + + +YY GR + + + C G FIEA
Sbjct: 222 VVYFFDSEAAQGLTTMKIKEHMFQWFNHYYFTCGRFRRSESGRPFIKCNDCGSRFIEAKC 401
Query: 110 DVSLVEFGETPLPPFPCFEELLYDVPGSEQVLDSPLLLIQVTRLKCGGFIMALRFNHTMS 169
+L E+ + +P ++ L+ ++ SP +L Q+T+ KCGG + L + H +
Sbjct: 402 KKTLDEW--LAMKDWPLYKLLVSQQVIGPELSFSPSVLFQLTQFKCGGISLGLSWAHVLG 575
Query: 170 DGAGLKQFLSALAEMA--QGASQPLTPP 195
D F+++ ++ G P P
Sbjct: 576 DPLSASDFINSWGQVMSNMGMKTPSNSP 659
>TC19494 weakly similar to UP|Q9SND9 (Q9SND9) Anthranilate
N-hydroxycinnamoyl/benzoyltransferase-like protein,
partial (4%)
Length = 420
Score = 45.4 bits (106), Expect = 2e-05
Identities = 38/125 (30%), Positives = 58/125 (46%)
Frame = +2
Query: 22 PASSTPHEVKLLSDIDDQDGLRFNIPFIMMYRHEPSMADKDPVQAIRQALSRTLVYYYPF 81
P +TP LS D Q L + + +Y S + V +R +LS+ L +YPF
Sbjct: 53 PYETTPTPTLSLSHCD-QLKLPNHGSQLFLYTPNDSPPLFNSVHTLRTSLSKVLTLFYPF 229
Query: 82 AGRLKEGPGRKLMVDCTGEGVMFIEANADVSLVEFGETPLPPFPCFEELLYDVPGSEQVL 141
AGRL G + + C +G A +DV L +FG+ L E+L+ + S +
Sbjct: 230 AGRLCWTHGGRFELLCNAKGAQLQGATSDVPLNDFGD--LADTHVAEQLVPRIDYSVPIE 403
Query: 142 DSPLL 146
+ PLL
Sbjct: 404 EVPLL 418
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.323 0.140 0.434
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,194,831
Number of Sequences: 28460
Number of extensions: 117532
Number of successful extensions: 617
Number of sequences better than 10.0: 70
Number of HSP's better than 10.0 without gapping: 591
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 595
length of query: 437
length of database: 4,897,600
effective HSP length: 93
effective length of query: 344
effective length of database: 2,250,820
effective search space: 774282080
effective search space used: 774282080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0175.13


