

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0175.11
(923 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
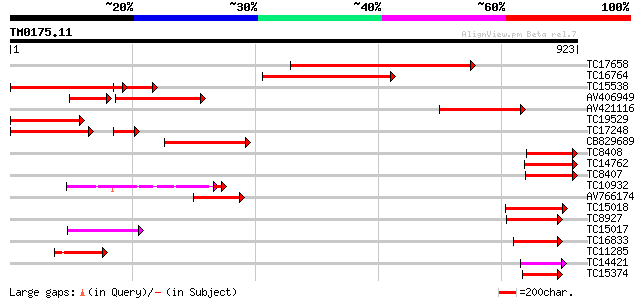
Score E
Sequences producing significant alignments: (bits) Value
TC17658 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp prot... 550 e-157
TC16764 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp prot... 397 e-111
TC15538 similar to UP|CLPA_PEA (P35100) ATP-dependent Clp protea... 293 1e-79
AV406949 286 1e-77
AV421116 271 3e-73
TC19529 similar to UP|CLPA_PEA (P35100) ATP-dependent Clp protea... 239 1e-63
TC17248 similar to UP|CLPA_PEA (P35100) ATP-dependent Clp protea... 227 6e-60
CB829689 185 2e-47
TC8408 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp prote... 162 2e-40
TC14762 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp prot... 151 4e-37
TC8407 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp prote... 146 1e-35
TC10932 homologue to UP|Q39889 (Q39889) Heat shock protein , par... 112 6e-31
AV766174 115 4e-26
TC15018 weakly similar to UP|ERD1_ARATH (P42762) ERD1 protein, c... 104 7e-23
TC8927 similar to UP|Q9LLI0 (Q9LLI0) ClpB, partial (12%) 80 1e-15
TC15017 67 1e-11
TC16833 similar to UP|Q9LF37 (Q9LF37) ClpB heat shock protein-li... 60 1e-09
TC11285 59 3e-09
TC14421 similar to UP|Q39889 (Q39889) Heat shock protein , parti... 56 3e-08
TC15374 similar to UP|Q9LF37 (Q9LF37) ClpB heat shock protein-li... 54 1e-07
>TC17658 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp protease
ATP-binding subunit clpA homolog, chloroplast precursor
, partial (33%)
Length = 908
Score = 550 bits (1416), Expect = e-157
Identities = 276/302 (91%), Positives = 295/302 (97%)
Frame = +1
Query: 457 LSYTDDALVAASQLSHQYISDRFLPDKAIDLIDEAGSRVRLRHAQLPEEARELDKEVRQI 516
L YTD++LVAA+QLS+QYISDRFLPDKAIDLIDEAGSRVRL+HAQLPEEARELDKEVR I
Sbjct: 1 LRYTDESLVAAAQLSYQYISDRFLPDKAIDLIDEAGSRVRLQHAQLPEEARELDKEVRLI 180
Query: 517 VKEKDEAVRNQDFEKAGELRDREMDLKTQISALIEKGKEMSKAESEADGAGPVVTEVDIQ 576
+KEK+E+VRNQDFEKAGELRD+EMDLK QISAL+EKGKEMSKAE+EA AGPVVTEVDIQ
Sbjct: 181 IKEKEESVRNQDFEKAGELRDKEMDLKAQISALVEKGKEMSKAETEAGDAGPVVTEVDIQ 360
Query: 577 HIVASWTGVPVDKVSSDESDRLLKMEETLHKRVIGQDEAVKAICRAIRRARVGLKNPNRP 636
HIV++WTG+PV+KVS+DESDRLLKME+TLHKR+IGQDEAVKAI RAIRRARVGLKNPNRP
Sbjct: 361 HIVSAWTGIPVEKVSTDESDRLLKMEDTLHKRIIGQDEAVKAISRAIRRARVGLKNPNRP 540
Query: 637 IASFIFSGPTGVGKSELAKTLASYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYT 696
IASFIFSGPTGVGKSELAK LA+YYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYT
Sbjct: 541 IASFIFSGPTGVGKSELAKALAAYYFGSEEAMIRLDMSEFMERHTVSKLIGSPPGYVGYT 720
Query: 697 EGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMT 756
EGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMT
Sbjct: 721 EGGQLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMT 900
Query: 757 SN 758
SN
Sbjct: 901 SN 906
>TC16764 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp protease
ATP-binding subunit clpA homolog, chloroplast precursor
, partial (24%)
Length = 653
Score = 397 bits (1021), Expect = e-111
Identities = 203/217 (93%), Positives = 210/217 (96%)
Frame = +3
Query: 412 EYRKHIEKDPALERRFQPVRVPEPTVDESIQILRGLRERYERHHKLSYTDDALVAASQLS 471
EYRKHIEKDPALERRFQPV+VPEPTVDE+IQIL+GLRERYE HHKL YTDDALVAA+ LS
Sbjct: 3 EYRKHIEKDPALERRFQPVKVPEPTVDETIQILKGLRERYEIHHKLRYTDDALVAAAHLS 182
Query: 472 HQYISDRFLPDKAIDLIDEAGSRVRLRHAQLPEEARELDKEVRQIVKEKDEAVRNQDFEK 531
+QYISDRFLPDKAIDLIDEAGSRVRLRHAQLPEEARELDKEVRQIVKEKDEAVRNQDFEK
Sbjct: 183 YQYISDRFLPDKAIDLIDEAGSRVRLRHAQLPEEARELDKEVRQIVKEKDEAVRNQDFEK 362
Query: 532 AGELRDREMDLKTQISALIEKGKEMSKAESEADGAGPVVTEVDIQHIVASWTGVPVDKVS 591
AGELRDREMDLKTQISALIEKGKEMSKAESEA GPVVTEVDIQHIVASWTG+PVDKVS
Sbjct: 363 AGELRDREMDLKTQISALIEKGKEMSKAESEAADGGPVVTEVDIQHIVASWTGIPVDKVS 542
Query: 592 SDESDRLLKMEETLHKRVIGQDEAVKAICRAIRRARV 628
+DESDRLLKMEETLHKRVIGQDEAVKAI RAIRRARV
Sbjct: 543 TDESDRLLKMEETLHKRVIGQDEAVKAISRAIRRARV 653
>TC15538 similar to UP|CLPA_PEA (P35100) ATP-dependent Clp protease
ATP-binding subunit clpA homolog, chloroplast precursor
, partial (21%)
Length = 658
Score = 293 bits (749), Expect = 1e-79
Identities = 150/192 (78%), Positives = 168/192 (87%)
Frame = +1
Query: 1 MSRVLAQSITIPGLVCGRSHGHNNRSTMSRRSLKMMSTLQAPALRMSGFSGLRTYNNLDT 60
M+RVLAQSI +P LV + HG++ S S+RS KMM LQ R++GFSGLRTYN LDT
Sbjct: 82 MARVLAQSINVPDLVARQRHGNHKGSGKSKRSAKMMCALQTSGFRLAGFSGLRTYNPLDT 261
Query: 61 MLRPGLDFRSKVFGVTTSRKARASRCIPKAMFERFTEKAIKVIMLSQEEARRLGHNFVGT 120
MLRPGLDF SKV T+SR+ +A+R +PKAMFERFTEKAIKVIML+QEEARRLGHNFVGT
Sbjct: 262 MLRPGLDFHSKVSIATSSRRGKATRGVPKAMFERFTEKAIKVIMLAQEEARRLGHNFVGT 441
Query: 121 EQILLGLVGEGTGIAARVLKAMGISLKDARVEVEKTIGRGSGFVAVEIPFTPRAKRVLEL 180
EQILLGL+GEGTGIAA+VLK+MGI+LKDARVEVEK IGRGSGFVAVEIPFTPRAKRVLEL
Sbjct: 442 EQILLGLIGEGTGIAAKVLKSMGINLKDARVEVEKIIGRGSGFVAVEIPFTPRAKRVLEL 621
Query: 181 SLEEARQLGHNY 192
SLEEARQLGHNY
Sbjct: 622 SLEEARQLGHNY 657
Score = 69.7 bits (169), Expect = 2e-12
Identities = 31/71 (43%), Positives = 52/71 (72%)
Frame = +1
Query: 170 FTPRAKRVLELSLEEARQLGHNYIGSEHLLLGLLREGEGVAARVLQDLGADPNNIRAQVI 229
FT +A +V+ L+ EEAR+LGHN++G+E +LLGL+ EG G+AA+VL+ +G + + R +V
Sbjct: 364 FTEKAIKVIMLAQEEARRLGHNFVGTEQILLGLIGEGTGIAAKVLKSMGINLKDARVEVE 543
Query: 230 RMVGESNNETA 240
+++G + A
Sbjct: 544 KIIGRGSGFVA 576
>AV406949
Length = 443
Score = 286 bits (731), Expect = 1e-77
Identities = 146/147 (99%), Positives = 146/147 (99%)
Frame = +1
Query: 172 PRAKRVLELSLEEARQLGHNYIGSEHLLLGLLREGEGVAARVLQDLGADPNNIRAQVIRM 231
PRAKRV ELSLEEARQLGHNYIGSEHLLLGLLREGEGVAARVLQDLGADPNNIRAQVIRM
Sbjct: 1 PRAKRVPELSLEEARQLGHNYIGSEHLLLGLLREGEGVAARVLQDLGADPNNIRAQVIRM 180
Query: 232 VGESNNETAGVAVGRGGSSNKMPTLEEYGTNLTKLANEGKLDPVVGRQQQIERVIQILGR 291
VGESNNETAGVAVGRGGSSNKMPTLEEYGTNLTKLANEGKLDPVVGRQQQIERVIQILGR
Sbjct: 181 VGESNNETAGVAVGRGGSSNKMPTLEEYGTNLTKLANEGKLDPVVGRQQQIERVIQILGR 360
Query: 292 RTKNNPCLVGEPGVGKTAIAEGLAQRI 318
RTKNNPCLVGEPGVGKTAIAEGLAQRI
Sbjct: 361 RTKNNPCLVGEPGVGKTAIAEGLAQRI 441
Score = 63.9 bits (154), Expect = 1e-10
Identities = 31/68 (45%), Positives = 47/68 (68%)
Frame = +1
Query: 98 KAIKVIMLSQEEARRLGHNFVGTEQILLGLVGEGTGIAARVLKAMGISLKDARVEVEKTI 157
+A +V LS EEAR+LGHN++G+E +LLGL+ EG G+AARVL+ +G + R +V + +
Sbjct: 4 RAKRVPELSLEEARQLGHNYIGSEHLLLGLLREGEGVAARVLQDLGADPNNIRAQVIRMV 183
Query: 158 GRGSGFVA 165
G + A
Sbjct: 184 GESNNETA 207
>AV421116
Length = 421
Score = 271 bits (694), Expect = 3e-73
Identities = 138/140 (98%), Positives = 140/140 (99%)
Frame = +1
Query: 700 QLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNV 759
+LTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNV
Sbjct: 1 RLTEAVRRRPYTVVLFDEIEKAHPDVFNMMLQILEDGRLTDSKGRTVDFKNTLLIMTSNV 180
Query: 760 GSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKL 819
GSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKL
Sbjct: 181 GSSVIEKGGRKIGFDLDYDEKDSSYNRIKSLVTEELKQYFRPEFLNRLDEMIVFRQLTKL 360
Query: 820 EVKEIADIMLKEVFDRLKTK 839
EVKEIADIMLKEVF+RLKTK
Sbjct: 361 EVKEIADIMLKEVFERLKTK 420
>TC19529 similar to UP|CLPA_PEA (P35100) ATP-dependent Clp protease
ATP-binding subunit clpA homolog, chloroplast precursor
, partial (8%)
Length = 560
Score = 239 bits (611), Expect = 1e-63
Identities = 121/121 (100%), Positives = 121/121 (100%)
Frame = -1
Query: 1 MSRVLAQSITIPGLVCGRSHGHNNRSTMSRRSLKMMSTLQAPALRMSGFSGLRTYNNLDT 60
MSRVLAQSITIPGLVCGRSHGHNNRSTMSRRSLKMMSTLQAPALRMSGFSGLRTYNNLDT
Sbjct: 365 MSRVLAQSITIPGLVCGRSHGHNNRSTMSRRSLKMMSTLQAPALRMSGFSGLRTYNNLDT 186
Query: 61 MLRPGLDFRSKVFGVTTSRKARASRCIPKAMFERFTEKAIKVIMLSQEEARRLGHNFVGT 120
MLRPGLDFRSKVFGVTTSRKARASRCIPKAMFERFTEKAIKVIMLSQEEARRLGHNFVGT
Sbjct: 185 MLRPGLDFRSKVFGVTTSRKARASRCIPKAMFERFTEKAIKVIMLSQEEARRLGHNFVGT 6
Query: 121 E 121
E
Sbjct: 5 E 3
Score = 38.1 bits (87), Expect = 0.006
Identities = 16/27 (59%), Positives = 23/27 (84%)
Frame = -1
Query: 170 FTPRAKRVLELSLEEARQLGHNYIGSE 196
FT +A +V+ LS EEAR+LGHN++G+E
Sbjct: 83 FTEKAIKVIMLSQEEARRLGHNFVGTE 3
>TC17248 similar to UP|CLPA_PEA (P35100) ATP-dependent Clp protease
ATP-binding subunit clpA homolog, chloroplast precursor
, partial (11%)
Length = 515
Score = 227 bits (579), Expect = 6e-60
Identities = 116/138 (84%), Positives = 128/138 (92%), Gaps = 2/138 (1%)
Frame = +3
Query: 1 MSRVLAQSITIPGLVCGRSHGHNNRSTMSRRSLKMM-STLQAPALRMSGFSGLRTYNNLD 59
MSRVLAQSI IPGLV GR+HG NNRST S+R +KMM +TLQAPALR+SGFSGLR++NNLD
Sbjct: 102 MSRVLAQSINIPGLVAGRTHGRNNRSTKSKRPVKMMYTTLQAPALRLSGFSGLRSFNNLD 281
Query: 60 TMLRPGLDFRSKVFGVTT-SRKARASRCIPKAMFERFTEKAIKVIMLSQEEARRLGHNFV 118
TMLRPGLDF SKVF TT SRKARASRC+PKAMFERFTEKAIKVIML+QEEARR+GHNFV
Sbjct: 282 TMLRPGLDFHSKVFATTTMSRKARASRCVPKAMFERFTEKAIKVIMLAQEEARRMGHNFV 461
Query: 119 GTEQILLGLVGEGTGIAA 136
GTEQ+LLGL+GEGTGIAA
Sbjct: 462 GTEQVLLGLIGEGTGIAA 515
Score = 53.9 bits (128), Expect = 1e-07
Identities = 23/42 (54%), Positives = 35/42 (82%)
Frame = +3
Query: 170 FTPRAKRVLELSLEEARQLGHNYIGSEHLLLGLLREGEGVAA 211
FT +A +V+ L+ EEAR++GHN++G+E +LLGL+ EG G+AA
Sbjct: 390 FTEKAIKVIMLAQEEARRMGHNFVGTEQVLLGLIGEGTGIAA 515
>CB829689
Length = 558
Score = 185 bits (470), Expect = 2e-47
Identities = 87/142 (61%), Positives = 113/142 (79%), Gaps = 1/142 (0%)
Frame = +1
Query: 252 KMPTLEEYGTNLTKLANEGKLDPVVGRQQQIERVIQILGRRTKNNPCLVGEPGVGKTAIA 311
K LE+YG +LT +A GKLDPV+GR +I R IQIL RRTKNNP L+GEPGVGKTAI+
Sbjct: 133 KYEALEKYGKDLTAMAKAGKLDPVIGRDDEIRRCIQILSRRTKNNPVLIGEPGVGKTAIS 312
Query: 312 EGLAQRIATGDVPETIEGKEVITLDMGLLVAGTKYRGEFEERLKKLMEEIKQND-NIILF 370
EGLAQRI GDVP+ + + +I+LDMG L+AG KYRGEFE+RLK +++E+ ++D +LF
Sbjct: 313 EGLAQRIVQGDVPQALMNRRLISLDMGALIAGAKYRGEFEDRLKAVLKEVTESDGQTVLF 492
Query: 371 IDEVHTLIGAGAAEGAIDAANI 392
IDE+HT++GAGA GA+DA N+
Sbjct: 493 IDEIHTVVGAGATNGAMDAGNL 558
>TC8408 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp protease
ATP-binding subunit clpA homolog, chloroplast precursor
, partial (9%)
Length = 498
Score = 162 bits (410), Expect = 2e-40
Identities = 83/83 (100%), Positives = 83/83 (100%)
Frame = +3
Query: 841 IDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDADS 900
IDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDADS
Sbjct: 3 IDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDADS 182
Query: 901 DGKVIVLNGSSGAPESLPEALPV 923
DGKVIVLNGSSGAPESLPEALPV
Sbjct: 183 DGKVIVLNGSSGAPESLPEALPV 251
>TC14762 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp protease
ATP-binding subunit clpA homolog, chloroplast precursor
, partial (9%)
Length = 588
Score = 151 bits (382), Expect = 4e-37
Identities = 76/85 (89%), Positives = 80/85 (93%)
Frame = +1
Query: 839 KEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDA 898
+EI+L VTERFRDRVV+EGYDPSYGARPLRRAIMRLLEDSMAEKMLA EIKEGDSVI+D
Sbjct: 1 QEIELPVTERFRDRVVDEGYDPSYGARPLRRAIMRLLEDSMAEKMLAREIKEGDSVIVDV 180
Query: 899 DSDGKVIVLNGSSGAPESLPEALPV 923
DSDG VIVLNGSSG PESLPEALPV
Sbjct: 181 DSDGNVIVLNGSSGTPESLPEALPV 255
>TC8407 homologue to UP|CLPA_PEA (P35100) ATP-dependent Clp protease
ATP-binding subunit clpA homolog, chloroplast precursor
, partial (8%)
Length = 603
Score = 146 bits (369), Expect = 1e-35
Identities = 76/86 (88%), Positives = 81/86 (93%), Gaps = 2/86 (2%)
Frame = +2
Query: 840 EIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDAD 899
EIDLSVTERF++RVV+EGY+PSYGARPLRRAIMRLLEDSMAEKMLA EIKEGDSVI+DAD
Sbjct: 2 EIDLSVTERFKERVVDEGYNPSYGARPLRRAIMRLLEDSMAEKMLAREIKEGDSVIVDAD 181
Query: 900 SDGKVIVLNGS--SGAPESLPEALPV 923
SDG VIVLNGS SGAPESLPE LPV
Sbjct: 182 SDGNVIVLNGSSDSGAPESLPEVLPV 259
>TC10932 homologue to UP|Q39889 (Q39889) Heat shock protein , partial (28%)
Length = 1001
Score = 112 bits (279), Expect(2) = 6e-31
Identities = 77/254 (30%), Positives = 129/254 (50%), Gaps = 6/254 (2%)
Frame = +3
Query: 93 ERFTEKAIKVIMLSQEEARRLGHNFVGTEQILLGLVGEGTGIAARVLKAMGISLKDARVE 152
++FT K + + + E A GH + + L+ + GI + + S +++
Sbjct: 210 DKFTHKTNEALAGAHELAMSSGHAQMTPLHLASTLISDPNGIFFQAIS--NSSGEESARA 383
Query: 153 VEKTIGRGSGFVAV------EIPFTPRAKRVLELSLEEARQLGHNYIGSEHLLLGLLREG 206
VE+ + + + EIP + + + + + G ++ + L+LG+L +
Sbjct: 384 VERVLNQALKKLPSQSPPPDEIPASTTLIKAIRRAQAAQKSRGDTHLAVDQLILGILEDS 563
Query: 207 EGVAARVLQDLGADPNNIRAQVIRMVGESNNETAGVAVGRGGSSNKMPTLEEYGTNLTKL 266
+ +L++ G +++++ ++ G+ G V L+ YG +L +
Sbjct: 564 Q--IGDLLKEAGVAAAKVKSELDKLRGK-----VGKKVESASGDTTFQALKTYGRDLVEQ 722
Query: 267 ANEGKLDPVVGRQQQIERVIQILGRRTKNNPCLVGEPGVGKTAIAEGLAQRIATGDVPET 326
A GKLDPV+GR ++I RV++IL RRTKNNP L+GEPGVGKTA+ EGLAQRI GDVP
Sbjct: 723 A--GKLDPVIGRDEEIRRVVRILSRRTKNNPVLIGEPGVGKTAVVEGLAQRIVRGDVPS- 893
Query: 327 IEGKEVITLDMGLL 340
I L +GLL
Sbjct: 894 ------IFLMLGLL 917
Score = 40.0 bits (92), Expect(2) = 6e-31
Identities = 18/22 (81%), Positives = 19/22 (85%)
Frame = +2
Query: 332 VITLDMGLLVAGTKYRGEFEER 353
+I LDMG LVAG KYRGEFEER
Sbjct: 911 LIALDMGALVAGAKYRGEFEER 976
>AV766174
Length = 416
Score = 115 bits (287), Expect = 4e-26
Identities = 52/84 (61%), Positives = 71/84 (83%), Gaps = 1/84 (1%)
Frame = -2
Query: 299 LVGEPGVGKTAIAEGLAQRIATGDVPETIEGKEVITLDMGLLVAGTKYRGEFEERLKKLM 358
L+GEPGVGKTAI+EGLAQRI GDVP+ + + +I+LDMG L+AG KYRGEFE+RLK ++
Sbjct: 412 LIGEPGVGKTAISEGLAQRIVQGDVPQALMNRRLISLDMGALIAGAKYRGEFEDRLKAVL 233
Query: 359 EEIKQND-NIILFIDEVHTLIGAG 381
+E+ ++D +LFIDE+HT++GAG
Sbjct: 232 KEVTESDGQTVLFIDEIHTVVGAG 161
>TC15018 weakly similar to UP|ERD1_ARATH (P42762) ERD1 protein, chloroplast
precursor, partial (10%)
Length = 598
Score = 104 bits (259), Expect = 7e-23
Identities = 48/102 (47%), Positives = 76/102 (74%)
Frame = +2
Query: 807 LDEMIVFRQLTKLEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARP 866
+DE++VFR L K ++ EI D++L++V R+ + IDL V+E ++ V +EGY+P+YGARP
Sbjct: 2 IDEIVVFRSLEKSQLLEILDVLLQDVKKRIMSLGIDLEVSESVKNLVCKEGYNPTYGARP 181
Query: 867 LRRAIMRLLEDSMAEKMLAGEIKEGDSVIIDADSDGKVIVLN 908
L+RAI L+ED ++E L G+ K+GD+V+ID D +G ++V N
Sbjct: 182 LKRAITSLIEDPLSEAFLCGKCKQGDTVLIDLDVNGNLLVTN 307
>TC8927 similar to UP|Q9LLI0 (Q9LLI0) ClpB, partial (12%)
Length = 619
Score = 80.1 bits (196), Expect = 1e-15
Identities = 38/91 (41%), Positives = 62/91 (67%)
Frame = +2
Query: 809 EMIVFRQLTKLEVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLR 868
E IVF+ L E+ +I ++ ++ V +RLK K+IDL T+ + + G+DP++GARP++
Sbjct: 2 EYIVFQPLDSKEISKIVELQMERVKNRLKQKKIDLHYTQEALELLSVLGFDPNFGARPVK 181
Query: 869 RAIMRLLEDSMAEKMLAGEIKEGDSVIIDAD 899
R I L+E+ +A +L G+ KE DS+I+DAD
Sbjct: 182 RVIQPLVENEIAMGVLRGDFKEDDSIIVDAD 274
>TC15017
Length = 1155
Score = 67.0 bits (162), Expect = 1e-11
Identities = 41/128 (32%), Positives = 71/128 (55%), Gaps = 3/128 (2%)
Frame = +2
Query: 94 RFTEKAIKVIMLSQEEARRLGHNFVGTEQILLGLVGEGTGIAARVLKAMGISLKDARVEV 153
+++ ++IK + + EAR+L + GTE +L+G++ EGT AA+ L+A GI+L R E
Sbjct: 263 KWSARSIKSFAMGELEARKLKYPTTGTEALLMGILVEGTSKAAKFLRANGITLFKVRDET 442
Query: 154 EKTIGRGS--GFVAVEIPFTPRAKRVLELSLEE-ARQLGHNYIGSEHLLLGLLREGEGVA 210
+ +G+ F P T A++ L+ +++E + G I HLLLG+ + E
Sbjct: 443 VELLGKSDMYFFSPEHPPLTEPARKALDWAIDEKLKSGGEGEINVAHLLLGIWSQKESAG 622
Query: 211 ARVLQDLG 218
++L LG
Sbjct: 623 QQILAGLG 646
>TC16833 similar to UP|Q9LF37 (Q9LF37) ClpB heat shock protein-like, partial
(10%)
Length = 510
Score = 60.5 bits (145), Expect = 1e-09
Identities = 26/80 (32%), Positives = 51/80 (63%)
Frame = +3
Query: 820 EVKEIADIMLKEVFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSM 879
++ I + L+ V R+ +++ + VTE + GYDP+YGARP++R I + +E+ +
Sbjct: 9 QISSIVRLQLERVQKRIADRKMKIQVTEAAIKLLGNLGYDPNYGARPVKRVIQQNVENEL 188
Query: 880 AEKMLAGEIKEGDSVIIDAD 899
A+ +L GE K+ D++++D +
Sbjct: 189 AKGILRGEFKDEDTILVDTE 248
>TC11285
Length = 456
Score = 58.9 bits (141), Expect = 3e-09
Identities = 32/87 (36%), Positives = 53/87 (60%)
Frame = +1
Query: 73 FGVTTSRKARASRCIPKAMFERFTEKAIKVIMLSQEEARRLGHNFVGTEQILLGLVGEGT 132
F + T+ +RA P A +++ KAIK + + EAR+L + GTE +L+G++ EGT
Sbjct: 193 FSLPTANPSRADS--PGAEIPKWSSKAIKSFAMGELEARKLKYPTTGTEALLMGVLIEGT 366
Query: 133 GIAARVLKAMGISLKDARVEVEKTIGR 159
+AA+ L+A G++L R E K +G+
Sbjct: 367 NVAAKFLRANGVTLFKVRDETVKLLGK 447
Score = 40.0 bits (92), Expect = 0.002
Identities = 23/71 (32%), Positives = 44/71 (61%), Gaps = 1/71 (1%)
Frame = +1
Query: 167 EIP-FTPRAKRVLELSLEEARQLGHNYIGSEHLLLGLLREGEGVAARVLQDLGADPNNIR 225
EIP ++ +A + + EAR+L + G+E LL+G+L EG VAA+ L+ G +R
Sbjct: 241 EIPKWSSKAIKSFAMGELEARKLKYPTTGTEALLMGVLIEGTNVAAKFLRANGVTLFKVR 420
Query: 226 AQVIRMVGESN 236
+ ++++G+++
Sbjct: 421 DETVKLLGKAD 453
>TC14421 similar to UP|Q39889 (Q39889) Heat shock protein , partial (14%)
Length = 731
Score = 55.8 bits (133), Expect = 3e-08
Identities = 28/75 (37%), Positives = 44/75 (58%)
Frame = +1
Query: 832 VFDRLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEG 891
V RL + I ++VT+ D ++ E YDP YGARP+RR + R + ++ ++ EI E
Sbjct: 1 VASRLAERGIAMAVTDAALDYILAESYDPVYGARPIRRWLERKVVTELSRMLIRDEIDEN 180
Query: 892 DSVIIDADSDGKVIV 906
+V IDA + G +V
Sbjct: 181 STVYIDAGTKGSELV 225
>TC15374 similar to UP|Q9LF37 (Q9LF37) ClpB heat shock protein-like, partial
(9%)
Length = 574
Score = 53.9 bits (128), Expect = 1e-07
Identities = 23/65 (35%), Positives = 44/65 (67%)
Frame = +2
Query: 835 RLKTKEIDLSVTERFRDRVVEEGYDPSYGARPLRRAIMRLLEDSMAEKMLAGEIKEGDSV 894
R+ +++ + VT+ + GYDP+YGARP++R I + +E+ +A+ +L GE K+ D++
Sbjct: 8 RIADRKMKIHVTDAAVQVLGSLGYDPNYGARPVKRVIQQNVENELAKGILRGEFKDEDTI 187
Query: 895 IIDAD 899
+ID +
Sbjct: 188 LIDTE 202
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.136 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,727,564
Number of Sequences: 28460
Number of extensions: 112836
Number of successful extensions: 609
Number of sequences better than 10.0: 70
Number of HSP's better than 10.0 without gapping: 591
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 600
length of query: 923
length of database: 4,897,600
effective HSP length: 99
effective length of query: 824
effective length of database: 2,080,060
effective search space: 1713969440
effective search space used: 1713969440
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0175.11


