

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0173b.8
(514 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
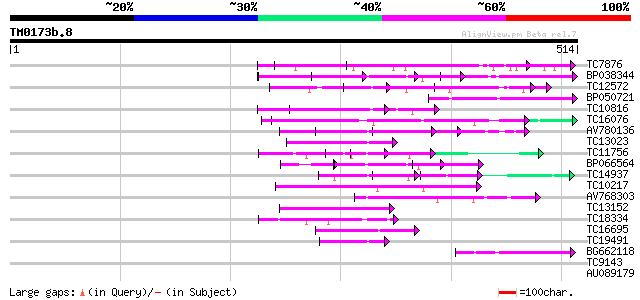
Score E
Sequences producing significant alignments: (bits) Value
TC7876 homologue to UP|GBLP_SOYBN (Q39836) Guanine nucleotide-bi... 101 3e-22
BP038344 77 7e-15
TC12572 similar to PIR|T46032|T46032 WD-40 repeat regulatory pro... 76 2e-14
BP050721 62 2e-10
TC10816 homologue to UP|CAE45585 (CAE45585) Coatomer alpha subun... 60 1e-09
TC16076 weakly similar to GB|AAG32022.1|11141605|AF277458 LEUNIG... 57 9e-09
AV780136 56 1e-08
TC13023 similar to UP|Q8L830 (Q8L830) WD40-repeat protein (Fragm... 54 8e-08
TC11756 weakly similar to GB|AAH01494.1|16306637|BC001494 U5 snR... 53 1e-07
BP066564 44 1e-06
TC14937 similar to UP|Q9FN19 (Q9FN19) Genomic DNA, chromosome 5,... 49 3e-06
TC10217 homologue to UP|AAQ54906 (AAQ54906) Cell cycle switch pr... 45 3e-05
AV768303 44 8e-05
TC13152 similar to UP|Q84XX6 (Q84XX6) Leunig (Fragment), partial... 43 1e-04
TC18334 weakly similar to UP|Q9LHN3 (Q9LHN3) Emb|CAB63739.1 (AT3... 42 2e-04
TC16695 homologue to UP|Q94KS2 (Q94KS2) TGF-beta receptor-intera... 42 3e-04
TC19491 similar to GB|AAP37692.1|30725340|BT008333 At4g29830 {Ar... 42 3e-04
BG662118 41 5e-04
TC9143 similar to UP|Q8H594 (Q8H594) OJ1634_B10.23 protein, part... 40 0.001
AU089179 40 0.001
>TC7876 homologue to UP|GBLP_SOYBN (Q39836) Guanine nucleotide-binding
protein beta subunit-like protein, complete
Length = 1379
Score = 101 bits (252), Expect = 3e-22
Identities = 81/301 (26%), Positives = 138/301 (44%), Gaps = 28/301 (9%)
Frame = +3
Query: 241 NSSRLITGGQDQSVKVW-----DTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNN 295
NS ++T +D+S+ +W D G L G V D+ ++ D + ++ S
Sbjct: 150 NSDMIVTASRDKSIILWHLTKEDKTYGVPRRRLTGHSHFVQDVVLSSDGQFALSGSWDGE 329
Query: 296 LYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIF 355
L +WDL +G GH V +V S + +R +VSA+ DRTIK+W+ + G C TI
Sbjct: 330 LRLWDLAAGTSARRFVGHTKDVLSVAFS-IDNRQIVSASRDRTIKLWNTL-GECKYTIQD 503
Query: 356 AS------NCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRN 409
+C S TI S D +++W++ + KL + +A HS V ++++S +
Sbjct: 504 NDAHSDWVSCVRFSPSTLQPTIVSASWDRTVKVWNLTNCKLRNTLAGHSGYVNTVAVSPD 683
Query: 410 GNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVH 469
G++ + G+D V L+D+ + S D G+ + + C SP N + + + S+
Sbjct: 684 GSLCASGGKDGVILLWDLAEGKRLYSL-DAGSIIHA----LCFSP-NRYWLCAATETSIK 845
Query: 470 IWSVHKNEIVSTLK--------------EHTSSVLCC---RWSGIGKPLASADKNGIVCI 512
IW + IV LK + V+ C WS G L S +G++ +
Sbjct: 846 IWDLESKSIVEDLKVDLKTEADATTGGNTNKKKVIYCTSLNWSADGSTLFSGYTDGVIRV 1025
Query: 513 W 513
W
Sbjct: 1026W 1028
Score = 88.6 bits (218), Expect = 2e-18
Identities = 68/267 (25%), Positives = 117/267 (43%), Gaps = 18/267 (6%)
Frame = +3
Query: 225 RLNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDN 284
RL H ++ + ++G D +++WD G+ G VL + + DN
Sbjct: 243 RLTGHSHFVQDVVLSSDGQFALSGSWDGELRLWDLAAGTSARRFVGHTKDVLSVAFSIDN 422
Query: 285 RSVIAASSSNNLYVWDLNSGRVRHTL---TGHKDKVCAVDVSKVSSR-HVVSAAYDRTIK 340
R +++AS + +W+ G ++T+ H D V V S + + +VSA++DRT+K
Sbjct: 423 RQIVSASRDRTIKLWN-TLGECKYTIQDNDAHSDWVSCVRFSPSTLQPTIVSASWDRTVK 599
Query: 341 VWDLMKGYCTNTIIFASN-CNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSL 399
VW+L NT+ S N + S DG SG DG + LWD+ GK L + A S+
Sbjct: 600 VWNLTNCKLRNTLAGHSGYVNTVAVSPDGSLCASGGKDGVILLWDLAEGKRLYSLDAGSI 779
Query: 400 AVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFR---------DTGNRVASNWSRS 450
+ ++ S N L + + ++D+ S + + TG
Sbjct: 780 -IHALCFSPN-RYWLCAATETSIKIWDLESKSIVEDLKVDLKTEADATTGGNTNKKKVIY 953
Query: 451 CI----SPDNSHVAAGSADGSVHIWSV 473
C S D S + +G DG + +W++
Sbjct: 954 CTSLNWSADGSTLFSGYTDGVIRVWAI 1034
Score = 79.3 bits (194), Expect = 1e-15
Identities = 61/218 (27%), Positives = 105/218 (47%), Gaps = 10/218 (4%)
Frame = +3
Query: 306 VRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMK-----GYCTNTIIFASN-C 359
+R T+ H D+V A+ +S +V+A+ D++I +W L K G + S+
Sbjct: 90 LRGTMRAHTDQVTAIATPIDNSDMIVTASRDKSIILWHLTKEDKTYGVPRRRLTGHSHFV 269
Query: 360 NALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRD 419
+ S DGQ SG DG LRLWD+ +G H+ V S++ S + ++++ RD
Sbjct: 270 QDVVLSSDGQFALSGSWDGELRLWDLAAGTSARRFVGHTKDVLSVAFSIDNRQIVSASRD 449
Query: 420 NVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCI--SPDNSH--VAAGSADGSVHIWSVHK 475
L++ +L C + N S+W SC+ SP + + S D +V +W++
Sbjct: 450 RTIKLWN--TLGEC-KYTIQDNDAHSDWV-SCVRFSPSTLQPTIVSASWDRTVKVWNLTN 617
Query: 476 NEIVSTLKEHTSSVLCCRWSGIGKPLASADKNGIVCIW 513
++ +TL H+ V S G AS K+G++ +W
Sbjct: 618 CKLRNTLAGHSGYVNTVAVSPDGSLCASGGKDGVILLW 731
>BP038344
Length = 525
Score = 77.0 bits (188), Expect = 7e-15
Identities = 43/147 (29%), Positives = 74/147 (50%), Gaps = 1/147 (0%)
Frame = +2
Query: 226 LNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNR 285
L AH+ A + F + + L + D+++ +W + T S+ L G + DL + D+
Sbjct: 86 LTAHDAAVACVKFSNDGTLLASASLDKTLIIWSSATLSLLHRLTGHSEGISDLAWSSDSH 265
Query: 286 SVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLM 345
+ +AS L +WD G TL GH V V+ + S ++VS ++D T++VW++
Sbjct: 266 YICSASDDRTLRIWDATGGDCVKTLRGHTHAVFCVNFNP-QSNYIVSGSFDETVRVWEVK 442
Query: 346 KGYCTNTII-FASNCNALCFSMDGQTI 371
G C +TII ++ F+ DG I
Sbjct: 443 TGRCIHTIIAHTMPVTSVHFNRDGTLI 523
Score = 76.6 bits (187), Expect = 9e-15
Identities = 39/142 (27%), Positives = 79/142 (55%), Gaps = 2/142 (1%)
Frame = +2
Query: 274 SVLDLTITHDNRSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSA 333
+V + ++D + +AS L +W + + H LTGH + + + S S ++ SA
Sbjct: 104 AVACVKFSNDGTLLASASLDKTLIIWSSATLSLLHRLTGHSEGISDLAWSS-DSHYICSA 280
Query: 334 AYDRTIKVWDLMKGYCTNTIIFASNCNALCFSMDGQTIY--SGHVDGNLRLWDIQSGKLL 391
+ DRT+++WD G C T+ ++ C + + Q+ Y SG D +R+W++++G+ +
Sbjct: 281 SDDRTLRIWDATGGDCVKTLRGHTHA-VFCVNFNPQSNYIVSGSFDETVRVWEVKTGRCI 457
Query: 392 SEVAAHSLAVTSISLSRNGNVV 413
+ AH++ VTS+ +R+G ++
Sbjct: 458 HTIIAHTMPVTSVHFNRDGTLI 523
Score = 51.6 bits (122), Expect = 3e-07
Identities = 22/100 (22%), Positives = 47/100 (47%)
Frame = +2
Query: 225 RLNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDN 284
RL H G + + + +S + + D+++++WD G L G +V + +
Sbjct: 209 RLTGHSEGISDLAWSSDSHYICSASDDRTLRIWDATGGDCVKTLRGHTHAVFCVNFNPQS 388
Query: 285 RSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSK 324
+++ S + VW++ +GR HT+ H V +V ++
Sbjct: 389 NYIVSGSFDETVRVWEVKTGRCIHTIIAHTMPVTSVHFNR 508
Score = 45.8 bits (107), Expect = 2e-05
Identities = 29/124 (23%), Positives = 57/124 (45%)
Frame = +2
Query: 391 LSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRS 450
L + AH AV + S +G ++ ++ D ++ +L + R TG+ + S
Sbjct: 77 LKTLTAHDAAVACVKFSNDGTLLASASLDKTLIIWSSATLSLLH--RLTGH--SEGISDL 244
Query: 451 CISPDNSHVAAGSADGSVHIWSVHKNEIVSTLKEHTSSVLCCRWSGIGKPLASADKNGIV 510
S D+ ++ + S D ++ IW + V TL+ HT +V C ++ + S + V
Sbjct: 245 AWSSDSHYICSASDDRTLRIWDATGGDCVKTLRGHTHAVFCVNFNPQSNYIVSGSFDETV 424
Query: 511 CIWK 514
+W+
Sbjct: 425 RVWE 436
>TC12572 similar to PIR|T46032|T46032 WD-40 repeat regulatory protein tup1
homolog - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (51%)
Length = 770
Score = 75.9 bits (185), Expect = 2e-14
Identities = 52/181 (28%), Positives = 90/181 (48%), Gaps = 7/181 (3%)
Frame = +3
Query: 303 SGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIFASN--CN 360
SG+ L H D V AVD ++ S +VS++YD ++WD G+C T+I N +
Sbjct: 6 SGKCLKVLPAHSDPVTAVDFNRDGSL-IVSSSYDGLCRIWDASTGHCMKTLIDDENPPVS 182
Query: 361 ALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAH---SLAVTSISLSRNGNVVLTSG 417
+ FS + + I G +D NLRLW+ +G+ L H ++S + NG V+
Sbjct: 183 FVKFSPNAKFILVGTLDNNLRLWNYSTGRFLKTYTGHVNSKYCISSTFSTTNGKYVVGGS 362
Query: 418 RDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSA--DGSVHIWSVHK 475
D+ L+++++ ++ + V S SC P + +A+G+ D +V IW+ K
Sbjct: 363 EDHGIYLWELQTRKIVQKLEGHSDTVV---SVSC-HPTENMIASGALGNDKTVKIWTQQK 530
Query: 476 N 476
+
Sbjct: 531 D 533
Score = 52.8 bits (125), Expect = 1e-07
Identities = 29/118 (24%), Positives = 54/118 (45%), Gaps = 7/118 (5%)
Frame = +3
Query: 236 ILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHG------CIGSVLDLTITHDNRSVIA 289
+ F N+ ++ G D ++++W+ +TG G CI S T + + V+
Sbjct: 186 VKFSPNAKFILVGTLDNNLRLWNYSTGRFLKTYTGHVNSKYCISSTFSTT---NGKYVVG 356
Query: 290 ASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSA-AYDRTIKVWDLMK 346
S + +Y+W+L + ++ L GH D V +V + A D+T+K+W K
Sbjct: 357 GSEDHGIYLWELQTRKIVQKLEGHSDTVVSVSCHPTENMIASGALGNDKTVKIWTQQK 530
Score = 48.5 bits (114), Expect = 3e-06
Identities = 26/106 (24%), Positives = 51/106 (47%)
Frame = +3
Query: 386 QSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVAS 445
+SGK L + AHS VT++ +R+G+++++S D + ++D + + D N S
Sbjct: 3 KSGKCLKVLPAHSDPVTAVDFNRDGSLIVSSSYDGLCRIWDASTGHCMKTLIDDENPPVS 182
Query: 446 NWSRSCISPDNSHVAAGSADGSVHIWSVHKNEIVSTLKEHTSSVLC 491
SP+ + G+ D ++ +W+ + T H +S C
Sbjct: 183 FVK---FSPNAKFILVGTLDNNLRLWNYSTGRFLKTYTGHVNSKYC 311
>BP050721
Length = 562
Score = 62.4 bits (150), Expect = 2e-10
Identities = 39/135 (28%), Positives = 68/135 (49%)
Frame = -1
Query: 380 LRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFRDT 439
+R +DI+ G+ +S+ V IS+S +GN +L D+ L D S E+ ++
Sbjct: 559 VRTFDIRIGREISDNFGQP--VNCISMSNDGNCILAGCLDSTVRLLDRTSGELLQEYKGH 386
Query: 440 GNRVASNWSRSCISPDNSHVAAGSADGSVHIWSVHKNEIVSTLKEHTSSVLCCRWSGIGK 499
N+ S C++ ++HVA GS DG ++ W + +VS+ + HTS V +
Sbjct: 385 TNK--SYKLDCCLTNTDAHVAGGSEDGFIYFWDLVDAYVVSSFRAHTSVVTSVSYHPKEN 212
Query: 500 PLASADKNGIVCIWK 514
+ SA +G + +WK
Sbjct: 211 CMLSASVDGTIRVWK 167
Score = 33.9 bits (76), Expect = 0.065
Identities = 21/93 (22%), Positives = 40/93 (42%), Gaps = 3/93 (3%)
Frame = -1
Query: 330 VVSAAYDRTIKVWDLMKGYCTNTIIFASNCNA---LCFSMDGQTIYSGHVDGNLRLWDIQ 386
+++ D T+++ D G +N + C + + G DG + WD+
Sbjct: 466 ILAGCLDSTVRLLDRTSGELLQEYKGHTNKSYKLDCCLTNTDAHVAGGSEDGFIYFWDLV 287
Query: 387 SGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRD 419
++S AH+ VTS+S N +L++ D
Sbjct: 286 DAYVVSSFRAHTSVVTSVSYHPKENCMLSASVD 188
>TC10816 homologue to UP|CAE45585 (CAE45585) Coatomer alpha subunit-like
protein, partial (20%)
Length = 979
Score = 59.7 bits (143), Expect = 1e-09
Identities = 32/121 (26%), Positives = 59/121 (48%), Gaps = 1/121 (0%)
Frame = +1
Query: 225 RLNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDN 284
+ + H+G + F + ++GG D +KVW+ L G + + + H++
Sbjct: 376 KFDEHDGPVRGVHFHHSQPLFVSGGDDYKIKVWNYKLHRCLFTLLGHLDYIRTVQFHHES 555
Query: 285 RSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKV-CAVDVSKVSSRHVVSAAYDRTIKVWD 343
+++AS + +W+ S LTGH V CA+ + VVSA+ D+T++VWD
Sbjct: 556 PWIVSASDDQTIRIWNWQSRTCISVLTGHNHYVMCALFHPREDL--VVSASLDQTVRVWD 729
Query: 344 L 344
+
Sbjct: 730 I 732
Score = 53.5 bits (127), Expect = 8e-08
Identities = 35/138 (25%), Positives = 60/138 (43%), Gaps = 2/138 (1%)
Frame = +1
Query: 254 VKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNSGRVRHTLTGH 313
+++WD G++ G V + H ++ + VW+ R TL GH
Sbjct: 337 IQLWDYRMGTLIDKFDEHDGPVRGVHFHHSQPLFVSGGDDYKIKVWNYKLHRCLFTLLGH 516
Query: 314 KDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIFASNCNALC--FSMDGQTI 371
D + V S +VSA+ D+TI++W+ C +++ N +C F +
Sbjct: 517 LDYIRTVQFHH-ESPWIVSASDDQTIRIWNWQSRTCI-SVLTGHNHYVMCALFHPREDLV 690
Query: 372 YSGHVDGNLRLWDIQSGK 389
S +D +R+WDI S K
Sbjct: 691 VSASLDQTVRVWDIGSLK 744
Score = 38.1 bits (87), Expect = 0.003
Identities = 33/148 (22%), Positives = 56/148 (37%), Gaps = 27/148 (18%)
Frame = +1
Query: 222 CKFRLNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTIT 281
C F L H ++ F S +++ DQ++++W+ + + S L G V+
Sbjct: 493 CLFTLLGHLDYIRTVQFHHESPWIVSASDDQTIRIWNWQSRTCISVLTGHNHYVMCALFH 672
Query: 282 HDNRSVIAASSSNNLYVWDLNSGR---------------------------VRHTLTGHK 314
V++AS + VWD+ S + V++ L GH
Sbjct: 673 PREDLVVSASLDQTVRVWDIGSLKRKNASPADDILRLSQMNTDLFGGVDAVVKYVLEGHD 852
Query: 315 DKVCAVDVSKVSSRHVVSAAYDRTIKVW 342
V +VSAA DR +K+W
Sbjct: 853 RGVNWASFHPALPL-IVSAADDRQVKLW 933
Score = 32.7 bits (73), Expect = 0.15
Identities = 28/137 (20%), Positives = 59/137 (42%), Gaps = 1/137 (0%)
Frame = +1
Query: 378 GNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFR 437
G ++LWD + G L+ + H V + + + ++ G D +++ + L C F
Sbjct: 331 GVIQLWDYRMGTLIDKFDEHDGPVRGVHFHHSQPLFVSGGDDYKIKVWNYK-LHRCL-FT 504
Query: 438 DTGNRVASNWSRSC-ISPDNSHVAAGSADGSVHIWSVHKNEIVSTLKEHTSSVLCCRWSG 496
G+ ++ R+ ++ + + S D ++ IW+ +S L H V+C +
Sbjct: 505 LLGHL---DYIRTVQFHHESPWIVSASDDQTIRIWNWQSRTCISVLTGHNHYVMCALFHP 675
Query: 497 IGKPLASADKNGIVCIW 513
+ SA + V +W
Sbjct: 676 REDLVVSASLDQTVRVW 726
>TC16076 weakly similar to GB|AAG32022.1|11141605|AF277458 LEUNIG
{Arabidopsis thaliana;} , partial (24%)
Length = 1048
Score = 56.6 bits (135), Expect = 9e-09
Identities = 56/279 (20%), Positives = 110/279 (39%), Gaps = 2/279 (0%)
Frame = +3
Query: 238 FEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLY 297
F + L + G D+ V +W+ +T S + D+ ++ + AS ++
Sbjct: 18 FSSDGKMLASAGDDKKVVLWNMDTLQTESTPEQHKSVISDVRFRPNSSQLATASIDKSVR 197
Query: 298 VWDL-NSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIFA 356
+WD N GH + ++D + I+ W++ CT +
Sbjct: 198 LWDAANPTYCVQEYNGHSSAIMSLDFHPKKTDIFCFCDSANEIRYWNITSSSCTR--VSK 371
Query: 357 SNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTS 416
+ + F + S D + ++D++S + + + H V SI NG+ L S
Sbjct: 372 GGSSQVRFQPRIGQVLSAASD*VVSIFDVESDRQIYTLQGHPEPVNSICWDVNGDF-LAS 548
Query: 417 GRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCI-SPDNSHVAAGSADGSVHIWSVHK 475
N+ ++ + S E +GN+ S C+ P S + S+ +W++
Sbjct: 549 VSPNLVKIWSLTSGECIQELSSSGNQFYS-----CVFHPSYSTLLVIGGFSSLELWNMAD 713
Query: 476 NEIVSTLKEHTSSVLCCRWSGIGKPLASADKNGIVCIWK 514
N+ + T+ H S + S + +ASA + V +WK
Sbjct: 714 NKSM-TISAHESVISALAQSPVTGMVASASHDNSVKLWK 827
Score = 55.8 bits (133), Expect = 2e-08
Identities = 57/252 (22%), Positives = 108/252 (42%), Gaps = 9/252 (3%)
Frame = +3
Query: 229 HEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVC-SNLHGCIGSVLDLTITHDNRSV 287
H+ + + F NSS+L T D+SV++WD + C +G +++ L +
Sbjct: 117 HKSVISDVRFRPNSSQLATASIDKSVRLWDAANPTYCVQEYNGHSSAIMSLDFHPKKTDI 296
Query: 288 IA-ASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSR--HVVSAAYDRTIKVWDL 344
S+N + W++ S G +V + R V+SAA D + ++D+
Sbjct: 297 FCFCDSANEIRYWNITSSSCTRVSKGGSSQV------RFQPRIGQVLSAASD*VVSIFDV 458
Query: 345 MKGYCTNTII-FASNCNALCFSMDGQTIYSGHVDGNL-RLWDIQSGKLLSEVAAHSLAVT 402
T+ N++C+ ++G + S V NL ++W + SG+ + E+++
Sbjct: 459 ESDRQIYTLQGHPEPVNSICWDVNGDFLAS--VSPNLVKIWSLTSGECIQELSSSGNQFY 632
Query: 403 SISLSRNGNVVLTSGRDN---VHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHV 459
S + + +L G + + N+ D +S+ + + S S SP V
Sbjct: 633 SCVFHPSYSTLLVIGGFSSLELWNMADNKSMTISAH--------ESVISALAQSPVTGMV 788
Query: 460 AAGSADGSVHIW 471
A+ S D SV +W
Sbjct: 789 ASASHDNSVKLW 824
Score = 26.9 bits (58), Expect = 8.0
Identities = 12/25 (48%), Positives = 15/25 (60%)
Frame = +3
Query: 489 VLCCRWSGIGKPLASADKNGIVCIW 513
V CC +S GK LASA + V +W
Sbjct: 3 VTCCHFSSDGKMLASAGDDKKVVLW 77
>AV780136
Length = 558
Score = 56.2 bits (134), Expect = 1e-08
Identities = 39/167 (23%), Positives = 75/167 (44%), Gaps = 1/167 (0%)
Frame = -1
Query: 245 LITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNSG 304
L++GG ++ ++VWD +GS L G ++ L + R ++ SS + + +WD+
Sbjct: 528 LVSGGTEKVIRVWDPRSGSKTLKLRGHADNIRALLLDSTGRYCLSGSSDSMIRLWDIGQP 349
Query: 305 RVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIFASNCNALCF 364
R HT H D V A+ S + HV S D ++ + DL ++++
Sbjct: 348 RCLHTYAVHTDSVWAL-ASTPTFSHVYSGGRDFSLYLTDLQTR--ESSLLCTGEHPIRQL 178
Query: 365 SMDGQTIYSGHVDGNLRLWDIQ-SGKLLSEVAAHSLAVTSISLSRNG 410
++ +I+ D ++ W + S + +S ++S SR G
Sbjct: 177 ALHDDSIWVASTDSSVHKWPAEGSNPQKIFLRGNSFLAGNLSFSRAG 37
Score = 53.9 bits (128), Expect = 6e-08
Identities = 45/143 (31%), Positives = 66/143 (45%), Gaps = 1/143 (0%)
Frame = -1
Query: 330 VVSAAYDRTIKVWDLMKGYCTNTII-FASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSG 388
+VS ++ I+VWD G T + A N AL G+ SG D +RLWDI
Sbjct: 528 LVSGGTEKVIRVWDPRSGSKTLKLRGHADNIRALLLDSTGRYCLSGSSDSMIRLWDIGQP 349
Query: 389 KLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWS 448
+ L A H+ +V +++ + + V + GRD L D+++ E SS TG
Sbjct: 348 RCLHTYAVHTDSVWALASTPTFSHVYSGGRDFSLYLTDLQTRE--SSLLCTGEHPI---- 187
Query: 449 RSCISPDNSHVAAGSADGSVHIW 471
R D+S + S D SVH W
Sbjct: 186 RQLALHDDS-IWVASTDSSVHKW 121
Score = 45.4 bits (106), Expect = 2e-05
Identities = 29/111 (26%), Positives = 54/111 (48%), Gaps = 1/111 (0%)
Frame = -1
Query: 278 LTITHDNRSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDR 337
L + +++ + + VWD SG L GH D + A+ + + R+ +S + D
Sbjct: 555 LAMNEGGTVLVSGGTEKVIRVWDPRSGSKTLKLRGHADNIRALLLDS-TGRYCLSGSSDS 379
Query: 338 TIKVWDLMKGYCTNT-IIFASNCNALCFSMDGQTIYSGHVDGNLRLWDIQS 387
I++WD+ + C +T + + AL + +YSG D +L L D+Q+
Sbjct: 378 MIRLWDIGQPRCLHTYAVHTDSVWALASTPTFSHVYSGGRDFSLYLTDLQT 226
Score = 33.9 bits (76), Expect = 0.065
Identities = 20/91 (21%), Positives = 42/91 (45%), Gaps = 4/91 (4%)
Frame = -1
Query: 403 SISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVAS----NWSRSCISPDNSH 458
+++++ G V+++ G + V ++D RS R + + + + R C+S
Sbjct: 558 ALAMNEGGTVLVSGGTEKVIRVWDPRSGSKTLKLRGHADNIRALLLDSTGRYCLS----- 394
Query: 459 VAAGSADGSVHIWSVHKNEIVSTLKEHTSSV 489
GS+D + +W + + + T HT SV
Sbjct: 393 ---GSSDSMIRLWDIGQPRCLHTYAVHTDSV 310
>TC13023 similar to UP|Q8L830 (Q8L830) WD40-repeat protein (Fragment),
partial (18%)
Length = 591
Score = 53.5 bits (127), Expect = 8e-08
Identities = 29/100 (29%), Positives = 52/100 (52%)
Frame = +1
Query: 252 QSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNSGRVRHTLT 311
+S+K+ D+ S+ S L G SV L ++ D+ + ++ S + VWDL++ + +
Sbjct: 223 ESIKIVDSANASIRSTLQGDSESVTALALSPDDNLLFSSGHSRQIRVWDLSTLKCVRSWK 402
Query: 312 GHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTN 351
GH+ V + S + + DR + VWD+ GYCT+
Sbjct: 403 GHEGPVMCMSCHP-SGGLLATGGADRKVLVWDVDGGYCTH 519
Score = 38.9 bits (89), Expect = 0.002
Identities = 23/121 (19%), Positives = 46/121 (38%)
Frame = +1
Query: 200 DGVVRRCEEGAEFFLDSNIPSTCKFRLNAHEGGCASILFEFNSSRLITGGQDQSVKVWDT 259
DG C G + + ++ + L ++ + + L + G + ++VWD
Sbjct: 193 DGSFIACACGESIKIVDSANASIRSTLQGDSESVTALALSPDDNLLFSSGHSRQIRVWDL 372
Query: 260 NTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCA 319
+T + G G V+ ++ + + + VWD++ G H GH V
Sbjct: 373 STLKCVRSWKGHEGPVMCMSCHPSGGLLATGGADRKVLVWDVDGGYCTHFFKGHGGVVSC 552
Query: 320 V 320
V
Sbjct: 553 V 555
Score = 30.8 bits (68), Expect = 0.55
Identities = 17/80 (21%), Positives = 35/80 (43%)
Frame = +1
Query: 434 SSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVHIWSVHKNEIVSTLKEHTSSVLCCR 493
+S R T + + + +SPD++ + + + +W + + V + K H V+C
Sbjct: 253 ASIRSTLQGDSESVTALALSPDDNLLFSSGHSRQIRVWDLSTLKCVRSWKGHEGPVMCMS 432
Query: 494 WSGIGKPLASADKNGIVCIW 513
G LA+ + V +W
Sbjct: 433 CHPSGGLLATGGADRKVLVW 492
>TC11756 weakly similar to GB|AAH01494.1|16306637|BC001494 U5 snRNP-specific
40 kDa protein (hPrp8-binding) {Homo sapiens;} , partial
(43%)
Length = 719
Score = 52.8 bits (125), Expect = 1e-07
Identities = 34/122 (27%), Positives = 57/122 (45%), Gaps = 3/122 (2%)
Frame = +1
Query: 226 LNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSN---LHGCIGSVLDLTITH 282
L H+ ++ F + + +G D+ + +W N C N L G +VLDL T
Sbjct: 292 LTGHQSVIYTMKFNPAGTVIASGSHDREIFLW--NVHGECKNFMVLKGHKNAVLDLHWTT 465
Query: 283 DNRSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVW 342
D +++AS + VWD+ +G+ + H V + S+ VVS + D T K+W
Sbjct: 466 DGTQIVSASPDKTVRVWDVETGKQVKKMVEHLSYVNSCCPSRRGPPLVVSGSDDGTAKLW 645
Query: 343 DL 344
D+
Sbjct: 646 DM 651
Score = 47.0 bits (110), Expect = 7e-06
Identities = 38/178 (21%), Positives = 70/178 (38%), Gaps = 3/178 (1%)
Frame = +1
Query: 310 LTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTIIFASNCNA---LCFSM 366
LTGH+ + + + + + S ++DR I +W++ G C N ++ + NA L ++
Sbjct: 292 LTGHQSVIYTMKFNPAGTV-IASGSHDREIFLWNV-HGECKNFMVLKGHKNAVLDLHWTT 465
Query: 367 DGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFD 426
DG I S D +R+WD+++GK + ++ H V S SR G ++ S
Sbjct: 466 DGTQIVSASPDKTVRVWDVETGKQVKKMVEHLSYVNSCCPSRRGPPLVVS---------- 615
Query: 427 VRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVHIWSVHKNEIVSTLKE 484
GS DG+ +W + + + T +
Sbjct: 616 -----------------------------------GSDDGTAKLWDMRQRGSIQTFPD 684
Score = 45.4 bits (106), Expect = 2e-05
Identities = 29/104 (27%), Positives = 55/104 (52%), Gaps = 4/104 (3%)
Frame = +1
Query: 287 VIAASSSNNLYVWDLNSGRVRH--TLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDL 344
+ + S +++W+++ G ++ L GHK+ V + + +VSA+ D+T++VWD+
Sbjct: 349 IASGSHDREIFLWNVH-GECKNFMVLKGHKNAVLDLHWT-TDGTQIVSASPDKTVRVWDV 522
Query: 345 MKGYCTNTII-FASNCNALCFSMDG-QTIYSGHVDGNLRLWDIQ 386
G ++ S N+ C S G + SG DG +LWD++
Sbjct: 523 ETGKQVKKMVEHLSYVNSCCPSRRGPPLVVSGSDDGTAKLWDMR 654
>BP066564
Length = 492
Score = 44.3 bits (103), Expect(2) = 1e-06
Identities = 26/103 (25%), Positives = 52/103 (50%), Gaps = 2/103 (1%)
Frame = +1
Query: 295 NLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYCTNTII 354
+L+ WD + ++ L GH V V + +S V++A++D T+K+WD+ C T+
Sbjct: 193 SLFWWDKQTTQLLEELKGHDGPVSCVRM--LSGERVLTASHDGTVKMWDVRTDRCVATVG 366
Query: 355 FASNCNALCFSMDGQ--TIYSGHVDGNLRLWDIQSGKLLSEVA 395
S+ LC D + + D +WDI++ + + +++
Sbjct: 367 RCSSA-VLCMEYDDNVGVLAAAGRDVVANIWDIRASRQMHKLS 492
Score = 43.9 bits (102), Expect = 6e-05
Identities = 18/64 (28%), Positives = 38/64 (59%)
Frame = +1
Query: 366 MDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLF 425
+ G+ + + DG +++WD+++ + ++ V S AV + N V+ +GRD V N++
Sbjct: 277 LSGERVLTASHDGTVKMWDVRTDRCVATVGRCSSAVLCMEYDDNVGVLAAAGRDVVANIW 456
Query: 426 DVRS 429
D+R+
Sbjct: 457 DIRA 468
Score = 36.6 bits (83), Expect = 0.010
Identities = 20/65 (30%), Positives = 33/65 (50%), Gaps = 1/65 (1%)
Frame = +1
Query: 450 SCISP-DNSHVAAGSADGSVHIWSVHKNEIVSTLKEHTSSVLCCRWSGIGKPLASADKNG 508
SC+ V S DG+V +W V + V+T+ +S+VLC + LA+A ++
Sbjct: 262 SCVRMLSGERVLTASHDGTVKMWDVRTDRCVATVGRCSSAVLCMEYDDNVGVLAAAGRDV 441
Query: 509 IVCIW 513
+ IW
Sbjct: 442 VANIW 456
Score = 24.6 bits (52), Expect(2) = 1e-06
Identities = 17/53 (32%), Positives = 23/53 (43%)
Frame = +3
Query: 246 ITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYV 298
I+G D SVK+WD + GS L T+ R++ A SS V
Sbjct: 48 ISGSTDCSVKIWDPSLR----------GSELRATLKGHTRTIRAISSDRGKVV 176
>TC14937 similar to UP|Q9FN19 (Q9FN19) Genomic DNA, chromosome 5, TAC
clone:K8K14 (AT5g67320/K8K14_4), partial (19%)
Length = 1089
Score = 48.5 bits (114), Expect = 3e-06
Identities = 24/93 (25%), Positives = 53/93 (56%), Gaps = 2/93 (2%)
Frame = +1
Query: 281 THDNRSVIAASSS--NNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRT 338
++ N+ ++ AS+S + + +WD+ G++ ++L GH+D V +V S + ++ S + D++
Sbjct: 31 SNPNKKLVLASASFDSTVKLWDVEVGKLIYSLNGHRDGVYSVAFSP-NGEYLASGSPDKS 207
Query: 339 IKVWDLMKGYCTNTIIFASNCNALCFSMDGQTI 371
I +W L +G T + +C++ +G I
Sbjct: 208 IHIWSLKEGKIVKTYTGSGGIFEVCWNKEGDKI 306
Score = 46.2 bits (108), Expect = 1e-05
Identities = 33/140 (23%), Positives = 54/140 (38%)
Frame = +1
Query: 373 SGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEV 432
S D ++LWD++ GKL+ + H RD V+++
Sbjct: 61 SASFDSTVKLWDVEVGKLIYSLNGH--------------------RDGVYSV-------- 156
Query: 433 CSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVHIWSVHKNEIVSTLKEHTSSVLCC 492
SP+ ++A+GS D S+HIWS+ + +IV T C
Sbjct: 157 ------------------AFSPNGEYLASGSPDKSIHIWSLKEGKIVKTYTGSGGIFEVC 282
Query: 493 RWSGIGKPLASADKNGIVCI 512
W+ G +A+ N VC+
Sbjct: 283 -WNKEGDKIAACFANNTVCV 339
Score = 42.0 bits (97), Expect = 2e-04
Identities = 24/104 (23%), Positives = 54/104 (51%), Gaps = 5/104 (4%)
Frame = +1
Query: 330 VVSAAYDRTIKVWDLMKGYCTNTIIFASNCN-----ALCFSMDGQTIYSGHVDGNLRLWD 384
+ SA++D T+K+WD+ G +I++ N + ++ FS +G+ + SG D ++ +W
Sbjct: 55 LASASFDSTVKLWDVEVG----KLIYSLNGHRDGVYSVAFSPNGEYLASGSPDKSIHIWS 222
Query: 385 IQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDNVHNLFDVR 428
++ GK++ S + + ++ G+ + +N + D R
Sbjct: 223 LKEGKIVKTYTG-SGGIFEVCWNKEGDKIAACFANNTVCVLDFR 351
Score = 40.0 bits (92), Expect = 0.001
Identities = 19/77 (24%), Positives = 34/77 (43%)
Frame = +1
Query: 224 FRLNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHD 283
+ LN H G S+ F N L +G D+S+ +W G + G G + ++ +
Sbjct: 118 YSLNGHRDGVYSVAFSPNGEYLASGSPDKSIHIWSLKEGKIVKTYTGS-GGIFEVCWNKE 294
Query: 284 NRSVIAASSSNNLYVWD 300
+ A ++N + V D
Sbjct: 295 GDKIAACFANNTVCVLD 345
Score = 33.1 bits (74), Expect = 0.11
Identities = 16/55 (29%), Positives = 28/55 (50%)
Frame = +1
Query: 459 VAAGSADGSVHIWSVHKNEIVSTLKEHTSSVLCCRWSGIGKPLASADKNGIVCIW 513
+A+ S D +V +W V +++ +L H V +S G+ LAS + + IW
Sbjct: 55 LASASFDSTVKLWDVEVGKLIYSLNGHRDGVYSVAFSPNGEYLASGSPDKSIHIW 219
>TC10217 homologue to UP|AAQ54906 (AAQ54906) Cell cycle switch protein
CCS52a, partial (45%)
Length = 640
Score = 45.1 bits (105), Expect = 3e-05
Identities = 42/191 (21%), Positives = 80/191 (40%), Gaps = 5/191 (2%)
Frame = +2
Query: 242 SSRLITGGQDQSVKVWDTNTGSV-CSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWD 300
SS L +GG+D+++ S L G V L ++DNR + + + N L+VW+
Sbjct: 23 SSLLSSGGRDKNIYQRXIRAQEXYVSKLSGHKSXVCGLKWSYDNRELASGGNDNRLFVWN 202
Query: 301 LNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVS--AAYDRTIKVWDLMKGYCTNTIIFASN 358
+S + H V A+ S + S DR I+ W+ + + S
Sbjct: 203 QHSTQPVLKYCEHTAAVKAIAWSPHLHGLLASGGGTADRCIRFWNTTTNSHLSCMDTGSQ 382
Query: 359 CNALCFSMDGQTIYS--GHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTS 416
L +S + + S G+ + +W + L+ + H+ V +++S +G ++T
Sbjct: 383 VCNLVWSKNVNELVSTHGYSQNQIIVWRYPTMSKLATLTGHTYRVLYLAISPDGQTIVTG 562
Query: 417 GRDNVHNLFDV 427
D ++V
Sbjct: 563 AGDETLXFWNV 595
Score = 35.4 bits (80), Expect = 0.022
Identities = 38/167 (22%), Positives = 68/167 (39%), Gaps = 2/167 (1%)
Frame = +2
Query: 335 YDRTIKVWDLMKGYCTNTIIFASNCNALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEV 394
Y R I+ + Y + S L +S D + + SG D L +W+ S + + +
Sbjct: 62 YQRXIRA---QEXYVSKLSGHKSXVCGLKWSYDNRELASGGNDNRLFVWNQHSTQPVLKY 232
Query: 395 AAHSLAVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISP 454
H+ AV +I+ S + + +L SG T +R W+ +
Sbjct: 233 CEHTAAVKAIAWSPHLHGLLASGGG-------------------TADRCIRFWNTTT--- 346
Query: 455 DNSHVAAGSADGSVH--IWSVHKNEIVSTLKEHTSSVLCCRWSGIGK 499
NSH++ V +WS + NE+VST + ++ R+ + K
Sbjct: 347 -NSHLSCMDTGSQVCNLVWSKNVNELVSTHGYSQNQIIVWRYPTMSK 484
Score = 35.0 bits (79), Expect = 0.029
Identities = 15/43 (34%), Positives = 23/43 (52%)
Frame = +2
Query: 453 SPDNSHVAAGSADGSVHIWSVHKNEIVSTLKEHTSSVLCCRWS 495
S DN +A+G D + +W+ H + V EHT++V WS
Sbjct: 143 SYDNRELASGGNDNRLFVWNQHSTQPVLKYCEHTAAVKAIAWS 271
Score = 34.7 bits (78), Expect = 0.038
Identities = 40/174 (22%), Positives = 74/174 (41%), Gaps = 10/174 (5%)
Frame = +2
Query: 310 LTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDL-----MKGYCTNT-----IIFASNC 359
L+GHK VC + S +R + S D + VW+ + YC +T I ++ +
Sbjct: 104 LSGHKSXVCGLKWS-YDNRELASGGNDNRLFVWNQHSTQPVLKYCEHTAAVKAIAWSPHL 280
Query: 360 NALCFSMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRD 419
+ L S G D +R W+ + LS + S V ++ S+N N ++++
Sbjct: 281 HGLLASGGGTA------DRCIRFWNTTTNSHLSCMDTGS-QVCNLVWSKNVNELVSTHGY 439
Query: 420 NVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVHIWSV 473
+ + + R + TG+ + ISPD + G+ D ++ W+V
Sbjct: 440 SQNQIIVWRYPTMSKLATLTGHTYRVLYL--AISPDGQTIVTGAGDETLXFWNV 595
Score = 31.2 bits (69), Expect = 0.42
Identities = 37/177 (20%), Positives = 73/177 (40%), Gaps = 16/177 (9%)
Frame = +2
Query: 225 RLNAHEGGCASILFEFNSSRLITGGQDQSVKVWD-----------TNTGSV-----CSNL 268
+L+ H+ + + +++ L +GG D + VW+ +T +V +L
Sbjct: 101 KLSGHKSXVCGLKWSYDNRELASGGNDNRLFVWNQHSTQPVLKYCEHTAAVKAIAWSPHL 280
Query: 269 HGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSR 328
HG + S +R + +++ N ++ +++G L K+ V S
Sbjct: 281 HGLLASGGGTA----DRCIRFWNTTTNSHLSCMDTGSQVCNLVWSKNVNELVSTHGYSQN 448
Query: 329 HVVSAAYDRTIKVWDLMKGYCTNTIIFASNCNALCFSMDGQTIYSGHVDGNLRLWDI 385
++ Y K+ L G+ T +++ L S DGQTI +G D L W++
Sbjct: 449 QIIVWRYPTMSKLATL-TGH-TYRVLY------LAISPDGQTIVTGAGDETLXFWNV 595
Score = 28.1 bits (61), Expect = 3.6
Identities = 16/59 (27%), Positives = 29/59 (49%), Gaps = 1/59 (1%)
Frame = +2
Query: 456 NSHVAAGSADGSVHIWSVHKNEI-VSTLKEHTSSVLCCRWSGIGKPLASADKNGIVCIW 513
+S +++G D +++ + E VS L H S V +WS + LAS + + +W
Sbjct: 23 SSLLSSGGRDKNIYQRXIRAQEXYVSKLSGHKSXVCGLKWSYDNRELASGGNDNRLFVW 199
>AV768303
Length = 550
Score = 43.5 bits (101), Expect = 8e-05
Identities = 39/185 (21%), Positives = 84/185 (45%), Gaps = 16/185 (8%)
Frame = +1
Query: 313 HKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGY-CTNTIIFASNCNALCFSMDGQTI 371
HKD + A+ + + + VSA D+T+K+W + + C T++ A+ S DG +
Sbjct: 13 HKDSIRAIRFA-ANGKLFVSAGDDKTVKIWSPPQSWRCIATVLSEKRVTAVAISNDGCYV 189
Query: 372 YSGHVDGNLRLWDIQ---SGKLLSEVAAHSLA-VTSISLSRNGNVVLTSGRD-------- 419
G + + D++ K + + +H + +TS+ S +G +L++ RD
Sbjct: 190 CFADKFGVVWVVDLEQPLGDKKPTPLLSHYCSIITSLEFSPDGRFILSADRDFKIRVTCF 369
Query: 420 ---NVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSADGSVHIWSVHKN 476
++ +++S C + + +A ++ C S + +GS D +V +W +
Sbjct: 370 PDKPLNGAHEIQSF--CLGHTEFVSCLAFVPAKEC---PQSLLLSGSGDSTVRLWDISSG 534
Query: 477 EIVST 481
++ T
Sbjct: 535 ALLDT 549
>TC13152 similar to UP|Q84XX6 (Q84XX6) Leunig (Fragment), partial (60%)
Length = 453
Score = 42.7 bits (99), Expect = 1e-04
Identities = 26/106 (24%), Positives = 44/106 (40%), Gaps = 1/106 (0%)
Frame = +2
Query: 245 LITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDL-NS 303
L +GG D+ +W T++ + L + D+ + + +S + VWD+ N
Sbjct: 5 LASGGHDKKAVLWYTDSLKQKATLEEHSALITDVRFSPSMPRLATSSFDKTVKVWDVDNP 184
Query: 304 GRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVWDLMKGYC 349
G T TGH V ++D + S D I+ W + G C
Sbjct: 185 GYSLRTFTGHSASVMSLDFHPNKEDLICSCDSDGEIRYWSINNGSC 322
Score = 38.1 bits (87), Expect = 0.003
Identities = 27/98 (27%), Positives = 39/98 (39%), Gaps = 3/98 (3%)
Frame = +2
Query: 223 KFRLNAHEGGCASILFEFNSSRLITGGQDQSVKVWDT-NTGSVCSNLHGCIGSVLDLTIT 281
K L H + F + RL T D++VKVWD N G G SV+ L
Sbjct: 65 KATLEEHSALITDVRFSPSMPRLATSSFDKTVKVWDVDNPGYSLRTFTGHSASVMSLDF- 241
Query: 282 HDNRS--VIAASSSNNLYVWDLNSGRVRHTLTGHKDKV 317
H N+ + + S + W +N+G G K+
Sbjct: 242 HPNKEDLICSCDSDGEIRYWSINNGSCARVSKGGTTKM 355
>TC18334 weakly similar to UP|Q9LHN3 (Q9LHN3) Emb|CAB63739.1
(AT3g18860/MCB22_3), partial (21%)
Length = 595
Score = 42.4 bits (98), Expect = 2e-04
Identities = 39/134 (29%), Positives = 59/134 (43%), Gaps = 7/134 (5%)
Frame = +2
Query: 226 LNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSN--LHGCIGSVLDLTITHD 283
L+ HE I N + T +D++V+ W S+ L G V L
Sbjct: 137 LHGHEDDVRGICVCGNDG-IATSSRDKTVRFWSPENRKFVSSKVLVGHSSFVGPLVWIPP 313
Query: 284 NRSV----IAASSSNNLY-VWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRT 338
N + +A+ + L VWDL++G HTL GH+ +V ++ VVSA+ D T
Sbjct: 314 NPELPQGGVASGGMDTLVLVWDLSTGEKAHTLKGHQLQVTSI---AFDDGDVVSASVDCT 484
Query: 339 IKVWDLMKGYCTNT 352
++ W G CT T
Sbjct: 485 LRRW--KNGQCTET 520
Score = 36.6 bits (83), Expect = 0.010
Identities = 34/140 (24%), Positives = 67/140 (47%), Gaps = 8/140 (5%)
Frame = +2
Query: 285 RSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKVW-- 342
RS + +SS + D ++R L GH+D V + V + + +++ D+T++ W
Sbjct: 62 RSFLFSSSRSLSQGHDFKEYQLRCELHGHEDDVRGICVC--GNDGIATSSRDKTVRFWSP 235
Query: 343 DLMKGYCTNTII-FASNCNALCF-----SMDGQTIYSGHVDGNLRLWDIQSGKLLSEVAA 396
+ K + ++ +S L + + + SG +D + +WD+ +G+ +
Sbjct: 236 ENRKFVSSKVLVGHSSFVGPLVWIPPNPELPQGGVASGGMDTLVLVWDLSTGEKAHTLKG 415
Query: 397 HSLAVTSISLSRNGNVVLTS 416
H L VTSI+ +G+VV S
Sbjct: 416 HQLQVTSIAFD-DGDVVSAS 472
>TC16695 homologue to UP|Q94KS2 (Q94KS2) TGF-beta receptor-interacting
protein 1, partial (46%)
Length = 531
Score = 41.6 bits (96), Expect = 3e-04
Identities = 26/94 (27%), Positives = 44/94 (46%)
Frame = +3
Query: 278 LTITHDNRSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDR 337
L D + + + +N VW ++G T GH V DVS+ SSR + +A D+
Sbjct: 123 LKYNRDGDLLFSCAKDHNPTVWFADNGERLGTYRGHNGAVWCCDVSRDSSRLITGSA-DQ 299
Query: 338 TIKVWDLMKGYCTNTIIFASNCNALCFSMDGQTI 371
T K+W++ G T F S ++ S+ + +
Sbjct: 300 TAKLWNVQTGQQLFTFNFDSPARSVDLSVGDKLV 401
Score = 32.7 bits (73), Expect = 0.15
Identities = 13/22 (59%), Positives = 17/22 (77%)
Frame = +3
Query: 241 NSSRLITGGQDQSVKVWDTNTG 262
+SSRLITG DQ+ K+W+ TG
Sbjct: 264 DSSRLITGSADQTAKLWNVQTG 329
Score = 31.6 bits (70), Expect = 0.32
Identities = 15/80 (18%), Positives = 34/80 (41%)
Frame = +3
Query: 226 LNAHEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNR 285
+ HE + + + L + +D + VW + G G G+V ++ D+
Sbjct: 93 MKGHERPLTFLKYNRDGDLLFSCAKDHNPTVWFADNGERLGTYRGHNGAVWCCDVSRDSS 272
Query: 286 SVIAASSSNNLYVWDLNSGR 305
+I S+ +W++ +G+
Sbjct: 273 RLITGSADQTAKLWNVQTGQ 332
Score = 31.2 bits (69), Expect = 0.42
Identities = 19/85 (22%), Positives = 42/85 (49%)
Frame = +3
Query: 397 HSLAVTSISLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDN 456
H +T + +R+G+++ + +D+ ++ + E ++R V W +S D+
Sbjct: 102 HERPLTFLKYNRDGDLLFSCAKDHNPTVWFADNGERLGTYRGHNGAV---WCCD-VSRDS 269
Query: 457 SHVAAGSADGSVHIWSVHKNEIVST 481
S + GSAD + +W+V + + T
Sbjct: 270 SRLITGSADQTAKLWNVQTGQQLFT 344
>TC19491 similar to GB|AAP37692.1|30725340|BT008333 At4g29830 {Arabidopsis
thaliana;}, partial (39%)
Length = 527
Score = 41.6 bits (96), Expect = 3e-04
Identities = 20/63 (31%), Positives = 35/63 (54%)
Frame = +1
Query: 282 HDNRSVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYDRTIKV 341
+D R + AS N++++D + T++GH V VDVS + + + DRT+++
Sbjct: 49 YDPRLLFTASDDGNVHMYDAEGKALVGTMSGHASWVLCVDVSP-DGGAIATGSSDRTVRL 225
Query: 342 WDL 344
WDL
Sbjct: 226 WDL 234
Score = 40.0 bits (92), Expect = 0.001
Identities = 27/119 (22%), Positives = 52/119 (43%), Gaps = 8/119 (6%)
Frame = +1
Query: 245 LITGGQDQSVKVWDTNTGSVCSNLHGCIGSVLDLTITHDNRSVIAASSSNNLYVWDLNSG 304
L T D +V ++D ++ + G VL + ++ D ++ SS + +WDL
Sbjct: 64 LFTASDDGNVHMYDAEGKALVGTMSGHASWVLCVDVSPDGGAIATGSSDRTVRLWDLAMR 243
Query: 305 RVRHTLTGHKDKVCAVDV-----SKVSSRHVVSAAYDRTIKVWDLMKG---YCTNTIIF 355
T++ H D+V V + V + S + D++I ++D +CT + F
Sbjct: 244 ASVQTMSNHTDQVWGVAFRPPGGTDVRGGRLASVSDDKSISLYDYS*S*FRHCTQCLYF 420
Score = 37.7 bits (86), Expect = 0.005
Identities = 18/57 (31%), Positives = 26/57 (45%)
Frame = +1
Query: 452 ISPDNSHVAAGSADGSVHIWSVHKNEIVSTLKEHTSSVLCCRWSGIGKPLASADKNG 508
+SPD +A GS+D +V +W + V T+ HT V W +P D G
Sbjct: 169 VSPDGGAIATGSSDRTVRLWDLAMRASVQTMSNHTDQV----WGVAFRPPGGTDVRG 327
Score = 35.0 bits (79), Expect = 0.029
Identities = 17/51 (33%), Positives = 26/51 (50%)
Frame = +1
Query: 463 SADGSVHIWSVHKNEIVSTLKEHTSSVLCCRWSGIGKPLASADKNGIVCIW 513
S DG+VH++ +V T+ H S VLC S G +A+ + V +W
Sbjct: 76 SDDGNVHMYDAEGKALVGTMSGHASWVLCVDVSPDGGAIATGSSDRTVRLW 228
>BG662118
Length = 421
Score = 40.8 bits (94), Expect = 5e-04
Identities = 27/109 (24%), Positives = 57/109 (51%)
Frame = +3
Query: 405 SLSRNGNVVLTSGRDNVHNLFDVRSLEVCSSFRDTGNRVASNWSRSCISPDNSHVAAGSA 464
+LS+N + V+++ V +LF++ + +V ++F ++ + DN+ +A G
Sbjct: 15 ALSKNDSYVMSACGGKV-SLFNMMTFKVMTTFMQPP---PASTFLAFHPQDNNIIAIGME 182
Query: 465 DGSVHIWSVHKNEIVSTLKEHTSSVLCCRWSGIGKPLASADKNGIVCIW 513
D ++HI++V +E+ S LK H + +S L S+ + +C+W
Sbjct: 183 DSTIHIYNVRVDEVKSKLKGHHKRITGLAFSTNLNILVSSGADAQLCVW 329
>TC9143 similar to UP|Q8H594 (Q8H594) OJ1634_B10.23 protein, partial (21%)
Length = 543
Score = 40.0 bits (92), Expect = 0.001
Identities = 22/70 (31%), Positives = 35/70 (49%), Gaps = 2/70 (2%)
Frame = +2
Query: 445 SNWSRSCISPDNS--HVAAGSADGSVHIWSVHKNEIVSTLKEHTSSVLCCRWSGIGKPLA 502
++W SC DNS H+ + S DG V +W + +S ++ H+ VLC W G +
Sbjct: 56 TSWVSSCKWHDNSWFHLLSASYDGKVMLWDLRTAWPLSVIESHSDKVLCADW-WKGDSVI 232
Query: 503 SADKNGIVCI 512
S + +CI
Sbjct: 233 SGGADSKLCI 262
Score = 29.3 bits (64), Expect = 1.6
Identities = 16/53 (30%), Positives = 28/53 (52%), Gaps = 1/53 (1%)
Frame = +2
Query: 329 HVVSAAYDRTIKVWDLMKGYCTNTIIFASNCNALCFS-MDGQTIYSGHVDGNL 380
H++SA+YD + +WDL + ++I + + LC G ++ SG D L
Sbjct: 101 HLLSASYDGKVMLWDLRTAW-PLSVIESHSDKVLCADWWKGDSVISGGADSKL 256
Score = 28.1 bits (61), Expect = 3.6
Identities = 18/57 (31%), Positives = 26/57 (45%), Gaps = 2/57 (3%)
Frame = +2
Query: 282 HDNR--SVIAASSSNNLYVWDLNSGRVRHTLTGHKDKVCAVDVSKVSSRHVVSAAYD 336
HDN +++AS + +WDL + + H DKV D K S V+S D
Sbjct: 83 HDNSWFHLLSASYDGKVMLWDLRTAWPLSVIESHSDKVLCADWWKGDS--VISGGAD 247
Score = 27.7 bits (60), Expect = 4.7
Identities = 15/50 (30%), Positives = 27/50 (54%)
Frame = +2
Query: 371 IYSGHVDGNLRLWDIQSGKLLSEVAAHSLAVTSISLSRNGNVVLTSGRDN 420
+ S DG + LWD+++ LS + +HS V + G+ V++ G D+
Sbjct: 104 LLSASYDGKVMLWDLRTAWPLSVIESHSDKVLCADWWK-GDSVISGGADS 250
>AU089179
Length = 340
Score = 39.7 bits (91), Expect = 0.001
Identities = 25/79 (31%), Positives = 40/79 (49%), Gaps = 3/79 (3%)
Frame = +3
Query: 229 HEGGCASILFEFNSSRLITGGQDQSVKVWDTNTGSVC---SNLHGCIGSVLDLTITHDNR 285
HEG ++ F + + L TGG+D S++V+ V S H SV DL + + +
Sbjct: 3 HEGQQLALAFNNDGTALATGGEDGSLRVFKWPXMKVILEHSTAHS--SSVKDLHFSSNGK 176
Query: 286 SVIAASSSNNLYVWDLNSG 304
+++ S VWDL+SG
Sbjct: 177 WIVSLGSGGPCRVWDLSSG 233
Score = 27.7 bits (60), Expect = 4.7
Identities = 19/61 (31%), Positives = 28/61 (45%), Gaps = 2/61 (3%)
Frame = +3
Query: 455 DNSHVAAGSADGSVHI--WSVHKNEIVSTLKEHTSSVLCCRWSGIGKPLASADKNGIVCI 512
D + +A G DGS+ + W K I+ H+SSV +S GK + S G +
Sbjct: 39 DGTALATGGEDGSLRVFKWPXMK-VILEHSTAHSSSVKDLHFSSNGKWIVSLGSGGPCRV 215
Query: 513 W 513
W
Sbjct: 216 W 218
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.129 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,818,456
Number of Sequences: 28460
Number of extensions: 123562
Number of successful extensions: 850
Number of sequences better than 10.0: 103
Number of HSP's better than 10.0 without gapping: 694
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 807
length of query: 514
length of database: 4,897,600
effective HSP length: 94
effective length of query: 420
effective length of database: 2,222,360
effective search space: 933391200
effective search space used: 933391200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0173b.8


