

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0172.11
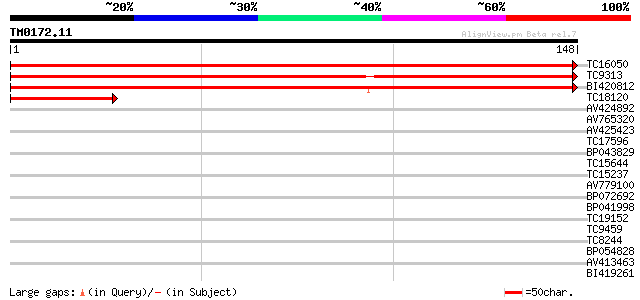
(148 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC16050 similar to UP|O82529 (O82529) Ribosomal protein L27a, pa... 281 3e-77
TC9313 homologue to UP|O82529 (O82529) Ribosomal protein L27a, p... 277 5e-76
BI420812 204 7e-54
TC18120 similar to UP|O82529 (O82529) Ribosomal protein L27a, pa... 47 1e-06
AV424892 28 0.85
AV765320 27 1.1
AV425423 27 1.9
TC17596 similar to UP|Q42768 (Q42768) Acetohydroxyacid synthase ... 27 1.9
BP043829 27 1.9
TC15644 weakly similar to UP|O80606 (O80606) At2g02870 protein (... 26 2.5
TC15237 similar to UP|AAQ24632 (AAQ24632) GPRP, partial (64%) 26 2.5
AV779100 25 4.2
BP072692 25 4.2
BP041998 25 4.2
TC19152 25 4.2
TC9459 similar to AAQ23982 (AAQ23982) Transcription factor Rap21... 25 5.5
TC8244 similar to PIR|S18343|DSPMN superoxide dismutase (Mn) pr... 25 5.5
BP054828 25 5.5
AV413463 25 7.2
BI419261 25 7.2
>TC16050 similar to UP|O82529 (O82529) Ribosomal protein L27a, partial (97%)
Length = 634
Score = 281 bits (720), Expect = 3e-77
Identities = 132/148 (89%), Positives = 138/148 (93%)
Frame = +3
Query: 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY
Sbjct: 51 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 230
Query: 61 FHKLRNQFFCPTVNIDKLWSLLPQEVKDQAANDKSKAPVIDVTNYGYFKVLGKGVLPANQ 120
FHKLRNQF CP VN++KLWSLLPQE KD+AA K APV+DVT YG+FKVLGKGVLP NQ
Sbjct: 231 FHKLRNQFHCPIVNVEKLWSLLPQEAKDKAAKSKGTAPVVDVTQYGFFKVLGKGVLPENQ 410
Query: 121 PVVVKAKLISKIAEKKIKENGGAVLLTA 148
P+VVKAKLISKIAEKKIKE GGAVLLTA
Sbjct: 411 PLVVKAKLISKIAEKKIKEAGGAVLLTA 494
>TC9313 homologue to UP|O82529 (O82529) Ribosomal protein L27a, partial
(94%)
Length = 624
Score = 277 bits (709), Expect = 5e-76
Identities = 131/148 (88%), Positives = 140/148 (94%)
Frame = +3
Query: 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY
Sbjct: 39 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 218
Query: 61 FHKLRNQFFCPTVNIDKLWSLLPQEVKDQAANDKSKAPVIDVTNYGYFKVLGKGVLPANQ 120
FHKLRNQF CP VNI+KLWSLLPQ+VKD+A+ D APV+DVT YGYFKVLGKGVLP+NQ
Sbjct: 219 FHKLRNQFHCPIVNIEKLWSLLPQDVKDKASKD--AAPVVDVTQYGYFKVLGKGVLPSNQ 392
Query: 121 PVVVKAKLISKIAEKKIKENGGAVLLTA 148
P+VVKAKL+SKIAEKKIKE GGAV+LTA
Sbjct: 393 PLVVKAKLVSKIAEKKIKEAGGAVVLTA 476
>BI420812
Length = 490
Score = 204 bits (518), Expect = 7e-54
Identities = 96/151 (63%), Positives = 113/151 (74%), Gaps = 3/151 (1%)
Frame = +1
Query: 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
M +R KNRKKRG+VS GHGR+GKHRKHPGGRGNAGG+HHHRI FDKYHPGYFGKVG+R+
Sbjct: 4 MPSRLGKNRKKRGNVSGGHGRVGKHRKHPGGRGNAGGLHHHRINFDKYHPGYFGKVGIRH 183
Query: 61 FHKLRNQFFCPTVNIDKLWSLLPQEVKDQAAN---DKSKAPVIDVTNYGYFKVLGKGVLP 117
+H RN FCP +NID LW LL +E +D+ N + PVIDVT YGY KVLGKG +P
Sbjct: 184 YHLKRNTEFCPVINIDHLWGLLSEEERDKVFNKQLPEGHVPVIDVTRYGYHKVLGKGKMP 363
Query: 118 ANQPVVVKAKLISKIAEKKIKENGGAVLLTA 148
P++VKAK ++ AEK IK GG LL A
Sbjct: 364 EGFPLIVKAKFFTEKAEKCIKAVGGVCLLVA 456
>TC18120 similar to UP|O82529 (O82529) Ribosomal protein L27a, partial (19%)
Length = 390
Score = 47.4 bits (111), Expect = 1e-06
Identities = 21/28 (75%), Positives = 22/28 (78%)
Frame = +3
Query: 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKH 28
MTT FKKN KKR HVS GHGRI K +KH
Sbjct: 297 MTTHFKKNMKKRSHVSTGHGRIEKLKKH 380
>AV424892
Length = 190
Score = 27.7 bits (60), Expect = 0.85
Identities = 12/31 (38%), Positives = 18/31 (57%)
Frame = -1
Query: 6 KKNRKKRGHVSAGHGRIGKHRKHPGGRGNAG 36
++ R++ GH +AG G G HR+ GG G
Sbjct: 100 RRRRRRHGHGAAGQGGAGAHRE--GGAARKG 14
>AV765320
Length = 572
Score = 27.3 bits (59), Expect = 1.1
Identities = 18/52 (34%), Positives = 23/52 (43%), Gaps = 2/52 (3%)
Frame = -3
Query: 19 HGRIGKHRKHPGGRGNAGGMHHH--RILFDKYHPGYFGKVGMRYFHKLRNQF 68
HG + +H +HPGG HHH R L G+ G+ Y H R F
Sbjct: 159 HGHV-RHCQHPGGGNGQWRDHHHWWRRLGGGGGEGWSGREDEPYLHGWRCVF 7
>AV425423
Length = 334
Score = 26.6 bits (57), Expect = 1.9
Identities = 17/46 (36%), Positives = 20/46 (42%)
Frame = -3
Query: 12 RGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVG 57
RG G + R+H GGRG A G DK H G+VG
Sbjct: 272 RGGQRQGSRECRRPRRHCGGRGGAAGGREGNSRADKGH----GEVG 147
>TC17596 similar to UP|Q42768 (Q42768) Acetohydroxyacid synthase , partial
(16%)
Length = 537
Score = 26.6 bits (57), Expect = 1.9
Identities = 11/35 (31%), Positives = 20/35 (56%)
Frame = +2
Query: 2 TTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAG 36
+T +++ R++ H+S + R+HPGG AG
Sbjct: 197 STTYRRPRRRDLHLSLPRR*ATQRRRHPGGGSGAG 301
>BP043829
Length = 497
Score = 26.6 bits (57), Expect = 1.9
Identities = 9/23 (39%), Positives = 14/23 (60%)
Frame = +3
Query: 6 KKNRKKRGHVSAGHGRIGKHRKH 28
+ +R ++GH GHG +HR H
Sbjct: 378 RSHRHRQGHGGGGHGHGHRHRHH 446
>TC15644 weakly similar to UP|O80606 (O80606) At2g02870 protein
(At2g02870/T17M13.4), partial (10%)
Length = 683
Score = 26.2 bits (56), Expect = 2.5
Identities = 10/25 (40%), Positives = 13/25 (52%)
Frame = -3
Query: 22 IGKHRKHPGGRGNAGGMHHHRILFD 46
+GK KH GNA HH I+ +
Sbjct: 207 LGKFTKHYSPNGNASASHHRTIIHE 133
>TC15237 similar to UP|AAQ24632 (AAQ24632) GPRP, partial (64%)
Length = 814
Score = 26.2 bits (56), Expect = 2.5
Identities = 16/45 (35%), Positives = 20/45 (43%)
Frame = +2
Query: 13 GHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVG 57
GH+ GHG+ +H K G+ G H H G FGK G
Sbjct: 512 GHM--GHGKFKQHGKFKHGKHGKFGKHKH---------GKFGKHG 613
>AV779100
Length = 561
Score = 25.4 bits (54), Expect = 4.2
Identities = 15/41 (36%), Positives = 18/41 (43%)
Frame = -2
Query: 11 KRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPG 51
+RG S GHG R G RG +GG L +Y G
Sbjct: 257 RRGSGSGGHGGRSWRR*GGGRRGGSGGQGRELGLACRYRRG 135
>BP072692
Length = 422
Score = 25.4 bits (54), Expect = 4.2
Identities = 11/25 (44%), Positives = 12/25 (48%)
Frame = -2
Query: 12 RGHVSAGHGRIGKHRKHPGGRGNAG 36
R V G G +G H GGRG G
Sbjct: 358 RRGVGGGEGEVGSHLGR*GGRGGRG 284
>BP041998
Length = 485
Score = 25.4 bits (54), Expect = 4.2
Identities = 10/31 (32%), Positives = 19/31 (61%)
Frame = -2
Query: 80 SLLPQEVKDQAANDKSKAPVIDVTNYGYFKV 110
SL+ + ++ ++ P++D TNY Y+KV
Sbjct: 343 SLIQVLMDNKEGGSVNRPPILDGTNYDYWKV 251
>TC19152
Length = 489
Score = 25.4 bits (54), Expect = 4.2
Identities = 11/25 (44%), Positives = 14/25 (56%)
Frame = -3
Query: 72 TVNIDKLWSLLPQEVKDQAANDKSK 96
T + + WSLL Q+ DQA K K
Sbjct: 343 TKQLPRNWSLLQQQTMDQAQTGKKK 269
>TC9459 similar to AAQ23982 (AAQ23982) Transcription factor Rap212,
partial (12%)
Length = 434
Score = 25.0 bits (53), Expect = 5.5
Identities = 11/24 (45%), Positives = 14/24 (57%)
Frame = +1
Query: 21 RIGKHRKHPGGRGNAGGMHHHRIL 44
RI +HR+HP R +H HR L
Sbjct: 70 RILRHRRHPHQRHRRLHLHRHRRL 141
>TC8244 similar to PIR|S18343|DSPMN superoxide dismutase (Mn) precursor -
garden pea {Pisum sativum;} , partial
(90%)
Length = 1168
Score = 25.0 bits (53), Expect = 5.5
Identities = 13/34 (38%), Positives = 17/34 (49%)
Frame = -2
Query: 57 GMRYFHKLRNQFFCPTVNIDKLWSLLPQEVKDQA 90
G R+F K+ P +N+ LWSL E D A
Sbjct: 408 GARFFQKMEWLT*PPPLNLIALWSLTTAEESDLA 307
>BP054828
Length = 476
Score = 25.0 bits (53), Expect = 5.5
Identities = 15/37 (40%), Positives = 20/37 (53%), Gaps = 1/37 (2%)
Frame = +2
Query: 10 KKRGHVSAGHGRIGK-HRKHPGGRGNAGGMHHHRILF 45
+K+G+ G R K R GRG+ GG+ H R LF
Sbjct: 239 RKKGNEGDGADRGQKLGRT*TDGRGHLGGVFHPRFLF 349
>AV413463
Length = 309
Score = 24.6 bits (52), Expect = 7.2
Identities = 12/28 (42%), Positives = 14/28 (49%)
Frame = +1
Query: 9 RKKRGHVSAGHGRIGKHRKHPGGRGNAG 36
R+ RGHV H +HP GRG G
Sbjct: 244 RRPRGHV---------HARHPSGRGADG 300
>BI419261
Length = 591
Score = 24.6 bits (52), Expect = 7.2
Identities = 11/32 (34%), Positives = 14/32 (43%)
Frame = +2
Query: 19 HGRIGKHRKHPGGRGNAGGMHHHRILFDKYHP 50
H + H +HP HHHR L + HP
Sbjct: 137 HAELLLHHRHPNP---PKPQHHHRPLSSRLHP 223
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.140 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,917,356
Number of Sequences: 28460
Number of extensions: 44786
Number of successful extensions: 241
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 237
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 238
length of query: 148
length of database: 4,897,600
effective HSP length: 82
effective length of query: 66
effective length of database: 2,563,880
effective search space: 169216080
effective search space used: 169216080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 51 (24.3 bits)
Lotus: description of TM0172.11


