

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
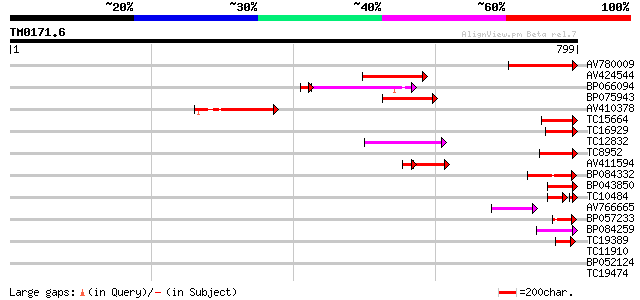
Query= TM0171.6
(799 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AV780009 129 1e-30
AV424544 103 8e-23
BP066094 92 2e-19
BP075943 92 4e-19
AV410378 89 2e-18
TC15664 weakly similar to UP|O81617 (O81617) F8M12.17 protein, p... 77 1e-14
TC16929 weakly similar to UP|Q9LFY6 (Q9LFY6) T7N9.5, partial (4%) 74 9e-14
TC12832 weakly similar to UP|POLX_TOBAC (P10978) Retrovirus-rela... 69 4e-12
TC8952 similar to UP|O82607 (O82607) T2L5.9 protein, partial (3%) 65 4e-11
AV411594 45 5e-07
BP084332 51 6e-07
BP043850 49 2e-06
TC10484 similar to UP|Q850H7 (Q850H7) Gag-pol polyprotein (Fragm... 39 3e-05
AV766665 42 3e-04
BP057233 42 4e-04
BP084259 41 6e-04
TC19389 weakly similar to UP|POLX_TOBAC (P10978) Retrovirus-rela... 41 8e-04
TC11910 similar to GB|CAA54498.1|602891|SCIILDNA YBL0422 {Saccha... 36 0.021
BP052124 36 0.021
TC19474 weakly similar to UP|Q9FJA1 (Q9FJA1) Similarity to retro... 32 0.51
>AV780009
Length = 529
Score = 129 bits (325), Expect = 1e-30
Identities = 60/97 (61%), Positives = 75/97 (76%)
Frame = -1
Query: 703 GCVDTRRSVTSYCFFIGNSLICWRSKKQQTISKSSSEAVYRALASATCEL*RLTYLLKDL 762
GC DTRRSVT Y F+G SLI WR+KKQ T+S+SSSEA YRALA+ CE+ L+YL + L
Sbjct: 418 GCPDTRRSVTGYSIFLGTSLISWRTKKQTTVSRSSSEAEYRALAATVCEVQWLSYLFQFL 239
Query: 763 QIEPVKPSVIYCDNQSALHIAANPVFHERTKHLEIEC 799
++ P ++CDNQSALHIA NP FHERTKH+E++C
Sbjct: 238 KLNVPLPVPLFCDNQSALHIAHNPTFHERTKHIELDC 128
>AV424544
Length = 276
Score = 103 bits (258), Expect = 8e-23
Identities = 53/91 (58%), Positives = 67/91 (73%)
Frame = +3
Query: 498 AKFTSLLVTCGYKQAHSDHSLFSKTQGQSFTILLIYVDDIILAGNFLDEFTRIKAALDNA 557
AK +S L GY Q+ DHSLF+K + SFT++L+YVDD+ILAGN L+E +K LD
Sbjct: 3 AKLSSYLHILGYIQSAHDHSLFTKFRDASFTVILVYVDDLILAGNDLNEIQCVKNKLDIQ 182
Query: 558 FKIKDLGVLKYFLGLEVSHSAKGISLCQRKY 588
F+IKDLG LKYFLGLEV+ S+ G+ L QRKY
Sbjct: 183 FRIKDLGTLKYFLGLEVARSSCGLFLSQRKY 275
>BP066094
Length = 532
Score = 91.7 bits (226), Expect(2) = 2e-19
Identities = 56/152 (36%), Positives = 87/152 (56%), Gaps = 4/152 (2%)
Frame = +2
Query: 426 AKLTIVRMVLALASVNNWHLHQLDVNNAFLLGDLYEDVYMKVPEGVSCV-DSGKVCKLHK 484
+K +R++++ + +N LHQ++V +AFL G + E+VY+ P G +S + KL K
Sbjct: 86 SKTEAIRLLISFSVNHNIILHQMNVKSAFLNGYISEEVYVHQPPGXEDEKNSDHIFKLKK 265
Query: 485 SSYGLKQASRQWYAKFTSLLVTCGYKQAHSDHSLFSKTQGQSFTILLIYVDDIILAG--- 541
S YGLKQA R WY + +S L+ + D +LF KT I+ IYVDDII
Sbjct: 266 SLYGLKQAPRAWYERLSSFLLENEXVRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANP 445
Query: 542 NFLDEFTRIKAALDNAFKIKDLGVLKYFLGLE 573
+ EF+ + A F+++ +G LKYFLG++
Sbjct: 446 SLCKEFSEMMQA---EFEMRMMGELKYFLGIQ 532
Score = 21.9 bits (45), Expect(2) = 2e-19
Identities = 9/18 (50%), Positives = 13/18 (72%)
Frame = +3
Query: 410 YNQIEGIYYFDTFSPTAK 427
Y+Q GI Y +TF+P A+
Sbjct: 39 YSQQ*GIDYTETFAPVAR 92
>BP075943
Length = 547
Score = 91.7 bits (226), Expect = 4e-19
Identities = 44/78 (56%), Positives = 57/78 (72%)
Frame = -3
Query: 526 SFTILLIYVDDIILAGNFLDEFTRIKAALDNAFKIKDLGVLKYFLGLEVSHSAKGISLCQ 585
SFT LL+YVDD++LAGN + E +K++L + F+IKDLG KYFLGLE++ S GI L Q
Sbjct: 533 SFTALLLYVDDVLLAGNDMHEIQLVKSSLHDQFRIKDLGEAKYFLGLEIARSTSGIVLNQ 354
Query: 586 RKYCLDLVHDSGVLGSKP 603
RKY L L+ DSG G +P
Sbjct: 353 RKYALQLISDSGHFGFQP 300
>AV410378
Length = 358
Score = 89.4 bits (220), Expect = 2e-18
Identities = 52/125 (41%), Positives = 76/125 (60%), Gaps = 7/125 (5%)
Frame = +1
Query: 261 PLPS-----PESPSTTSVSDHTRRPTRPRHQPSHLRNYVLHTVSSSCKASQ--TSSGIKY 313
PLPS PE + +S+ RRP PS+L++Y T+++S S+ +S+GI Y
Sbjct: 1 PLPSETLSNPEEGNVQRISNRVRRP------PSYLQDYHC-TLAASTTVSKVPSSAGISY 159
Query: 314 PISNYMSYSNLSIPHHAYAMSLSLDSEPHTYAEASKHKCWVDAMNLEISALEANGTWSLV 373
P+S +SY L+ + A+ M+++ SEP Y+EA KH+CW AM+ EI ALE N TW LV
Sbjct: 160 PLSKVISYHKLTPSYRAFIMNITTVSEPTRYSEAVKHECWRKAMDQEIEALERNHTWILV 339
Query: 374 PLPPN 378
PP+
Sbjct: 340 DKPPD 354
>TC15664 weakly similar to UP|O81617 (O81617) F8M12.17 protein, partial (4%)
Length = 670
Score = 76.6 bits (187), Expect = 1e-14
Identities = 33/50 (66%), Positives = 43/50 (86%)
Frame = +1
Query: 750 CEL*RLTYLLKDLQIEPVKPSVIYCDNQSALHIAANPVFHERTKHLEIEC 799
CEL* LTY+L+DL++ + PS++YCD+QSA HIA N VFHERTKHL+I+C
Sbjct: 1 CEL*WLTYILQDLRVPFISPSLLYCDSQSARHIATNAVFHERTKHLDIDC 150
>TC16929 weakly similar to UP|Q9LFY6 (Q9LFY6) T7N9.5, partial (4%)
Length = 553
Score = 73.9 bits (180), Expect = 9e-14
Identities = 31/45 (68%), Positives = 40/45 (88%)
Frame = +1
Query: 755 LTYLLKDLQIEPVKPSVIYCDNQSALHIAANPVFHERTKHLEIEC 799
LTYLL+DL++ +P+++YCDN SA HIAANPVFHERTKH+EI+C
Sbjct: 7 LTYLLQDLKVPFEQPALVYCDNNSARHIAANPVFHERTKHIEIDC 141
>TC12832 weakly similar to UP|POLX_TOBAC (P10978) Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
; Reverse transcriptase ; Endonuclease] , partial (9%)
Length = 747
Score = 68.6 bits (166), Expect = 4e-12
Identities = 40/119 (33%), Positives = 61/119 (50%), Gaps = 3/119 (2%)
Frame = +2
Query: 500 FTSLLVTCGYKQAHSDHSLFSKT-QGQSFTILLIYVDDIILAGNFLDEFTRIKAALDNAF 558
F S +++ GY + SDH + K F ILL+YVDD+++ G D +KA L F
Sbjct: 2 FDSFIMSLGYNRLSSDHCTYHKRFDDNDFIILLLYVDDMLVVGPNKDRVQELKAQLAREF 181
Query: 559 KIKDLGVLKYFLGLEVSHSAKG--ISLCQRKYCLDLVHDSGVLGSKPVSTPLDPSSRLS 615
+KDLG LG+++ K I L Q+ Y ++ + P+STPL + +LS
Sbjct: 182 DMKDLGPANKILGMQIHRDRKDRRIWLSQKNYLQKVLRRFNMQDCNPISTPLPVNYKLS 358
>TC8952 similar to UP|O82607 (O82607) T2L5.9 protein, partial (3%)
Length = 550
Score = 65.1 bits (157), Expect = 4e-11
Identities = 31/53 (58%), Positives = 40/53 (74%)
Frame = +3
Query: 747 SATCEL*RLTYLLKDLQIEPVKPSVIYCDNQSALHIAANPVFHERTKHLEIEC 799
S CE LTY L DL+I + VIYCDN+SALH+AAN VFH+RT+++EI+C
Sbjct: 9 SHPCEALWLTYALADLRIASLLLVVIYCDNRSALHLAANSVFHKRTENIEIDC 167
>AV411594
Length = 244
Score = 44.7 bits (104), Expect(2) = 5e-07
Identities = 23/51 (45%), Positives = 31/51 (60%)
Frame = +2
Query: 569 FLGLEVSHSAKGISLCQRKYCLDLVHDSGVLGSKPVSTPLDPSSRLSQDGG 619
FL ++ S KGI L +RKY L L+ D+ +G KP PLDPS +L+ G
Sbjct: 50 FLRT*IAKSKKGILLSRRKYALQLLDDTRNMGCKPPPFPLDPSIKLNSTDG 202
Score = 26.6 bits (57), Expect(2) = 5e-07
Identities = 12/20 (60%), Positives = 15/20 (75%)
Frame = +3
Query: 554 LDNAFKIKDLGVLKYFLGLE 573
L +FK+K L +KYFLGLE
Sbjct: 6 LAKSFKLKVLDDMKYFLGLE 65
>BP084332
Length = 368
Score = 51.2 bits (121), Expect = 6e-07
Identities = 25/70 (35%), Positives = 47/70 (66%), Gaps = 1/70 (1%)
Frame = +2
Query: 730 QQTISKSSSEAVYRALASATCEL*RLTYLLKDLQIEPVKPSV-IYCDNQSALHIAANPVF 788
Q TI+ S++EA Y + A + ++ + + L+D QI ++ ++ IYCDN +A+ ++ NP+
Sbjct: 32 QSTIALSTAEAEYISAAICSTQMLWMKHQLEDYQI--LESNIPIYCDNTAAISLSKNPIL 205
Query: 789 HERTKHLEIE 798
H R KH+E++
Sbjct: 206 HSRAKHIEVK 235
>BP043850
Length = 515
Score = 49.3 bits (116), Expect = 2e-06
Identities = 20/42 (47%), Positives = 31/42 (73%)
Frame = -1
Query: 758 LLKDLQIEPVKPSVIYCDNQSALHIAANPVFHERTKHLEIEC 799
LL +L +P+ ++ DN SA+ IAANPV+HE T+H+E++C
Sbjct: 491 LLSELGFLQSQPTPLHADNTSAIQIAANPVYHEWTRHIEVDC 366
>TC10484 similar to UP|Q850H7 (Q850H7) Gag-pol polyprotein (Fragment),
partial (20%)
Length = 479
Score = 38.5 bits (88), Expect(2) = 3e-05
Identities = 14/29 (48%), Positives = 23/29 (79%)
Frame = +3
Query: 758 LLKDLQIEPVKPSVIYCDNQSALHIAANP 786
+L ++ IE + P ++CDNQ+ALHI++NP
Sbjct: 15 ILSEMGIERISPMPLWCDNQAALHISSNP 101
Score = 26.6 bits (57), Expect(2) = 3e-05
Identities = 8/11 (72%), Positives = 11/11 (99%)
Frame = +2
Query: 789 HERTKHLEIEC 799
HERTKH+E++C
Sbjct: 113 HERTKHIEVDC 145
>AV766665
Length = 601
Score = 42.4 bits (98), Expect = 3e-04
Identities = 24/66 (36%), Positives = 36/66 (54%)
Frame = +3
Query: 679 SKKRVIFPKEFCSTIVGV**CRLGGCVDTRRSVTSYCFFIGNSLICWRSKKQQTISKSSS 738
S++ +F KE S++ G G + R S Y F+ +L+ WRSK+Q I++SS
Sbjct: 399 SRRGPLFQKEGKSSMDGFTYADYLGSIVDRLSTMGYYMFLSGNLVTWRSKQQNIIARSSG 578
Query: 739 EAVYRA 744
EA RA
Sbjct: 579 EAELRA 596
>BP057233
Length = 473
Score = 42.0 bits (97), Expect = 4e-04
Identities = 18/33 (54%), Positives = 26/33 (78%)
Frame = -2
Query: 766 PVKPSVIYCDNQSALHIAANPVFHERTKHLEIE 798
P KP +++CDN SA +A+NPV H R+KH+EI+
Sbjct: 460 PRKP-ILWCDNLSAKALASNPVLHARSKHIEID 365
>BP084259
Length = 385
Score = 41.2 bits (95), Expect = 6e-04
Identities = 20/57 (35%), Positives = 30/57 (52%)
Frame = -3
Query: 743 RALASATCEL*RLTYLLKDLQIEPVKPSVIYCDNQSALHIAANPVFHERTKHLEIEC 799
R + S E L LLK+LQ K I+ +N +A ++ N VFH KH+ ++C
Sbjct: 179 RTIGSTAAEFLWLQQLLKELQFSLPKVPCIFSNNINATYVCLNLVFHSHMKHVALDC 9
>TC19389 weakly similar to UP|POLX_TOBAC (P10978) Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
; Reverse transcriptase ; Endonuclease] , partial (6%)
Length = 498
Score = 40.8 bits (94), Expect = 8e-04
Identities = 13/28 (46%), Positives = 22/28 (78%)
Frame = +2
Query: 770 SVIYCDNQSALHIAANPVFHERTKHLEI 797
+V+YCDNQ A+++ N FH R+KH+++
Sbjct: 167 TVLYCDNQGAIYLGQNSTFHSRSKHIDV 250
>TC11910 similar to GB|CAA54498.1|602891|SCIILDNA YBL0422 {Saccharomyces
cerevisiae;} , partial (6%)
Length = 638
Score = 36.2 bits (82), Expect = 0.021
Identities = 21/58 (36%), Positives = 32/58 (54%)
Frame = +1
Query: 230 LPETSVPASSSNSLDDLSVSIPSADATSVQVPLPSPESPSTTSVSDHTRRPTRPRHQP 287
L +S+P SSS+S S S+ + +S +P PSP PS + R P++ RH+P
Sbjct: 22 LSSSSLPPSSSSSSSS-STSLHLSPCSSHSLPQPSPPPPSKSHFPSLPRFPSKCRHKP 192
>BP052124
Length = 467
Score = 36.2 bits (82), Expect = 0.021
Identities = 13/27 (48%), Positives = 18/27 (66%)
Frame = -2
Query: 773 YCDNQSALHIAANPVFHERTKHLEIEC 799
+CDN SAL +A P+ H R H E++C
Sbjct: 454 FCDNNSALTLAPRPI*HSRPVHFEVDC 374
>TC19474 weakly similar to UP|Q9FJA1 (Q9FJA1) Similarity to retroelement pol
polyprotein, partial (3%)
Length = 517
Score = 31.6 bits (70), Expect = 0.51
Identities = 12/32 (37%), Positives = 24/32 (74%)
Frame = -1
Query: 718 IGNSLICWRSKKQQTISKSSSEAVYRALASAT 749
+ ++LI W+SK+ +++S +EA +R +A+AT
Sbjct: 517 LSDNLISWKSKETIIVARSRAEAEFRVMATAT 422
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.338 0.145 0.471
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,039,676
Number of Sequences: 28460
Number of extensions: 231618
Number of successful extensions: 2612
Number of sequences better than 10.0: 103
Number of HSP's better than 10.0 without gapping: 2536
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2590
length of query: 799
length of database: 4,897,600
effective HSP length: 98
effective length of query: 701
effective length of database: 2,108,520
effective search space: 1478072520
effective search space used: 1478072520
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0171.6


