

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
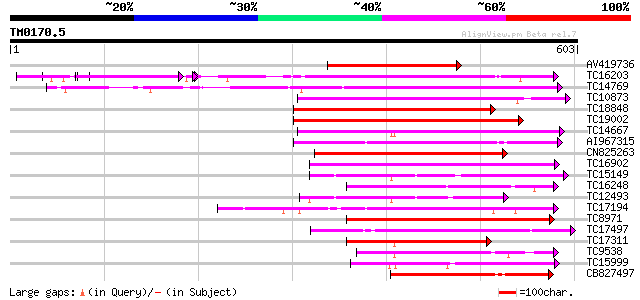
Query= TM0170.5
(603 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AV419736 291 3e-79
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 261 2e-70
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 223 7e-59
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 203 7e-53
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 201 4e-52
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 199 1e-51
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 191 4e-49
AI967315 189 1e-48
CN825263 187 5e-48
TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-... 184 3e-47
TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like pro... 171 3e-43
TC16248 similar to UP|BAC87845 (BAC87845) Leucine-rich repeat re... 163 8e-41
TC12493 similar to UP|Q9LP77 (Q9LP77) T1N15.9, partial (35%) 163 8e-41
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 163 8e-41
TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P... 157 6e-39
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 154 5e-38
TC17311 similar to UP|Q8K2Y2 (Q8K2Y2) Receptor-interacting serin... 152 2e-37
TC9538 similar to UP|RLK5_ARATH (P47735) Receptor-like protein k... 152 2e-37
TC15999 similar to UP|Q9LLT5 (Q9LLT5) Receptor-like protein kina... 149 1e-36
CB827497 146 1e-35
>AV419736
Length = 427
Score = 291 bits (744), Expect = 3e-79
Identities = 142/142 (100%), Positives = 142/142 (100%)
Frame = +2
Query: 339 TFAVKRIDRSREGCDQVFERELEILGSIKHINLVNLRGYCRLPSARLLIYDYLAIGSLDD 398
TFAVKRIDRSREGCDQVFERELEILGSIKHINLVNLRGYCRLPSARLLIYDYLAIGSLDD
Sbjct: 2 TFAVKRIDRSREGCDQVFERELEILGSIKHINLVNLRGYCRLPSARLLIYDYLAIGSLDD 181
Query: 399 LLHENTEQPLNWNDRLNIALGSARGLAYLHHECCPKIVHRDIKSSNILLNENMEPHISDF 458
LLHENTEQPLNWNDRLNIALGSARGLAYLHHECCPKIVHRDIKSSNILLNENMEPHISDF
Sbjct: 182 LLHENTEQPLNWNDRLNIALGSARGLAYLHHECCPKIVHRDIKSSNILLNENMEPHISDF 361
Query: 459 GLAKLLVDEDAHVTTVVAGTFG 480
GLAKLLVDEDAHVTTVVAGTFG
Sbjct: 362 GLAKLLVDEDAHVTTVVAGTFG 427
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 261 bits (667), Expect = 2e-70
Identities = 177/526 (33%), Positives = 281/526 (52%), Gaps = 15/526 (2%)
Frame = +2
Query: 73 VRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQ 132
++S++L ++ G I + ++ L ++ + N+L G IP IT+ L A+ L N
Sbjct: 1562 LQSLSLDANEFIGEIPGGVFEIPMLTKVNISGNNLTGPIPTTITHRASLTAVDLSRNNLA 1741
Query: 133 GGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIPDIGVLSTF 192
G +P + NL L+IL+LS N G +P + + L L+LS+N F+G +P G F
Sbjct: 1742 GEVPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSLTTLDLSSNNFTGTVPTGGQFLVF 1921
Query: 193 QKN-SFIGNLDLCGRQIQKPCRTSFGFPVVIPHAESDEA----AVPTKRSSSHYMKVVLI 247
+ +F GN +LC PH S + ++ R+ + ++ ++I
Sbjct: 1922 NYDKTFAGNPNLC-----------------FPHRASCPSVLYDSLRKTRAKTARVRAIVI 2050
Query: 248 G-AMTTLGLLLLVTLSFLWIRLLSKKERAVMRYTDVKKQVDPEASTKLITFHGDLPYTSS 306
G A+ T LL+ VT+ + R L + + + KL F L +
Sbjct: 2051 GIALATAVLLVAVTVHVVRKRRLHRAQ-----------------AWKLTAFQR-LEIKAE 2176
Query: 307 EIIEKLESLEEEDIVGSGGFGTVYRMVMNDCGTFAVKRIDRSREGC-DQVFERELEILGS 365
+++E L+EE+I+G GG G VYR M + A+KR+ G D F E+E LG
Sbjct: 2177 DVVE---CLKEENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETLGK 2347
Query: 366 IKHINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHENTEQPLNWNDRLNIALGSARGLA 425
I+H N++ L GY LL+Y+Y+ GSL + LH L W R IA+ +ARGL
Sbjct: 2348 IRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKGGHLRWEMRYKIAVEAARGLC 2527
Query: 426 YLHHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLVDEDAHVT-TVVAGTFGYLAP 484
Y+HH+C P I+HRD+KS+NILL+ + E H++DFGLAK L D A + + +AG++GY+AP
Sbjct: 2528 YMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAP 2707
Query: 485 EYLQSGRATEKSDVYSFGVLLLELVTGKRPTDPSFANRGLNVVGWMN-TLQKENRLED-- 541
EY + + EKSDVYSFGV+LLEL+ G++P F + G+++VGW+N T+ + ++ D
Sbjct: 2708 EYAYTLKVDEKSDVYSFGVVLLELIIGRKPVG-EFGD-GVDIVGWVNKTMSELSQPSDTA 2881
Query: 542 ----VVDRRCTDADAGTLEVILELAARCTDANADDRPSMNQVLQLL 583
VVD R + ++ + +A C RP+M +V+ +L
Sbjct: 2882 LVLAVVDPRLSGYPLTSVIHMFNIAMMCVKEMGPARPTMREVVHML 3019
Score = 90.5 bits (223), Expect = 7e-19
Identities = 62/201 (30%), Positives = 107/201 (52%), Gaps = 9/201 (4%)
Frame = +2
Query: 8 WVFILIFTTVFTPSSLALTQ--DGLTLLEIKGALNDTK---NVLSNWQEFDES---PCAW 59
++ +L FT ++ ++ + D LL++K ++ K + L +W+ F S C++
Sbjct: 128 YLLVLCFTLIWFRWTVVYSSFSDLDALLKLKESMKGAKAKHHALEDWK-FSTSLSAHCSF 304
Query: 60 TGITCHPGDGEQRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCT 119
+G+TC D RV ++N+ L G + P IG L +L+ L + N+L +P+++ + T
Sbjct: 305 SGVTC---DQNLRVVALNVTLVPLFGHLPPEIGLLEKLENLTISMNNLTDQLPSDLASLT 475
Query: 120 ELRALYLRANYFQGGIPSDIG-NLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNF 178
L+ L + N F G P +I + L LD NSF G +P + +L L+ L+L+ N+
Sbjct: 476 SLKVLNISHNLFSGQFPGNITVGMTELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGNY 655
Query: 179 FSGEIPDIGVLSTFQKNSFIG 199
FSG IP+ S FQ F+G
Sbjct: 656 FSGTIPE--SYSEFQSLEFLG 712
Score = 74.3 bits (181), Expect = 5e-14
Identities = 39/114 (34%), Positives = 63/114 (55%)
Frame = +2
Query: 71 QRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANY 130
+ +R + + L G I PS+G L++L L + N+L G IP E+++ L +L L N
Sbjct: 839 ENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMMSLMSLDLSIND 1018
Query: 131 FQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIP 184
G IP L L +++ N F+G++PS +G LP+L+ L + N FS +P
Sbjct: 1019LTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLP 1180
Score = 64.7 bits (156), Expect = 4e-11
Identities = 36/115 (31%), Positives = 63/115 (54%), Gaps = 1/115 (0%)
Frame = +2
Query: 71 QRVRSINLPYSQLGGIISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLR-AN 129
++++ ++L + G I S + L+ L L+ NSL G +P + L+ L+L +N
Sbjct: 620 EKLKYLHLAGNYFSGTIPESYSEFQSLEFLGLNANSLTGRVPESLAKLKTLKELHLGYSN 799
Query: 130 YFQGGIPSDIGNLPFLNILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIP 184
++GGIP G++ L +L++++ + G IP SLG L L L + N +G IP
Sbjct: 800 AYEGGIPPAFGSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIP 964
Score = 59.3 bits (142), Expect = 2e-09
Identities = 39/149 (26%), Positives = 65/149 (43%)
Frame = +2
Query: 36 KGALNDTKNVLSNWQEFDESPCAWTGITCHPGDGEQRVRSINLPYSQLGGIISPSIGKLS 95
+G + + N + + + C TG ++ S+ + + L G I P + +
Sbjct: 806 EGGIPPAFGSMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFVQMNNLTGTIPPELSSMM 985
Query: 96 RLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIPSDIGNLPFLNILDLSSNSF 155
L L L N L G IP + L + N F+G +PS IG+LP L L + N+F
Sbjct: 986 SLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNF 1165
Query: 156 KGAIPSSLGRLPHLQVLNLSTNFFSGEIP 184
+P +LG +++ N +G IP
Sbjct: 1166SFVLPHNLGGNGRFLYFDVTKNHLTGLIP 1252
Score = 57.8 bits (138), Expect = 5e-09
Identities = 33/122 (27%), Positives = 59/122 (48%), Gaps = 6/122 (4%)
Frame = +2
Query: 86 IISPSIGKLSRLQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIPSDIGNLPFL 145
++ ++G R + +N L G+IP ++ L+ + N+F+G IP IG L
Sbjct: 1172 VLPHNLGGNGRFLYFDVTKNHLTGLIPPDLCKSGRLKTFIITDNFFRGPIPKGIGECRSL 1351
Query: 146 NILDLSSNSFKGAIPSSLGRLPHLQVLNLSTNFFSGEIPDI------GVLSTFQKNSFIG 199
+ +++N G +P + +LP + + LS N +GE+P + G L T N F G
Sbjct: 1352 TKIRVANNFLDGPVPPGVFQLPSVTITELSNNRLNGELPSVISGESLGTL-TLSNNLFTG 1528
Query: 200 NL 201
+
Sbjct: 1529 KI 1534
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 223 bits (568), Expect = 7e-59
Identities = 177/567 (31%), Positives = 275/567 (48%), Gaps = 18/567 (3%)
Frame = +3
Query: 40 NDTKNVLSNWQEFDESPCA---WTGITCHPGDGEQRVRSINLPYSQLGGIISPSIGKLSR 96
N L +W PC W GI C +G + ++L S L G+I SI +++
Sbjct: 1500 NSGNRALESWSG---DPCILLPWKGIACDGSNGSSVITKLDLSSSNLKGLIPSSIAEMTN 1670
Query: 97 LQRLALHQNSLHGIIPNEITNCTELRALYLRANYFQGGIPSDIGNLPFLNIL---DLSSN 153
L+ L + NS F G +PS P ++L DLS N
Sbjct: 1671 LETLNISHNS------------------------FDGSVPS----FPLSSLLISVDLSYN 1766
Query: 154 SFKGAIPSSLGRLPHLQVLNLSTN-FFSGEIPDIGVLSTFQKNSFIGNLDLCGRQIQKPC 212
G +P S+ +LPHL+ L N S E P NS + N D GR
Sbjct: 1767 DLMGKLPESIVKLPHLKSLYFGCNEHMSPEDP-------ANMNSSLINTDY-GR------ 1904
Query: 213 RTSFGFPVVIPHAESDEAAVPTKRSSSHYMKVVLIGAMTTLGLLLLVTLSFLWIRLLSKK 272
K S + +V++IGA+T LL+ + L++ +K
Sbjct: 1905 ---------------------CKGKESRFGQVIVIGAITCGSLLITLAFGVLFVCRYRQK 2021
Query: 273 ERAVMRYTDVKKQVDPEASTKLITFHGDLPYTSSEI----IEKLESLEE--EDIVGSGGF 326
+ K ++ L + D S I +E +E E + ++G GGF
Sbjct: 2022 LIPWEGFAGKKYPMETNIIFSLPS-KDDFFIKSVSIQAFTLEYIEVATERYKTLIGEGGF 2198
Query: 327 GTVYRMVMNDCGTFAVK-RIDRSREGCDQVFERELEILGSIKHINLVNLRGYCRLPSARL 385
G+VYR +ND AVK R S +G + F+ EL +L +I+H NLV L GYC ++
Sbjct: 2199 GSVYRGTLNDGQEVAVKVRSSTSTQGTRE-FDNELNLLSAIQHENLVPLLGYCNESDQQI 2375
Query: 386 LIYDYLAIGSLDDLLHEN--TEQPLNWNDRLNIALGSARGLAYLHHECCPKIVHRDIKSS 443
L+Y +++ GSL D L+ + L+W RL+IALG+ARGLAYLH ++HRDIKSS
Sbjct: 2376 LVYPFMSNGSLQDRLYGEPAKRKILDWPTRLSIALGAARGLAYLHTFPGRSVIHRDIKSS 2555
Query: 444 NILLNENMEPHISDFGLAKLLVDE-DAHVTTVVAGTFGYLAPEYLQSGRATEKSDVYSFG 502
NILL+ +M ++DFG +K E D++V+ V GT GYL PEY ++ + +EKSDV+SFG
Sbjct: 2556 NILLDHSMCAKVADFGFSKYAPQEGDSYVSLEVRGTAGYLDPEYYKTQQLSEKSDVFSFG 2735
Query: 503 VLLLELVTGKRPTDPSFANRGLNVVGWMNTLQKENRLEDVVDRRCTDA-DAGTLEVILEL 561
V+LLE+V+G+ P + ++V W + ++++++VD A + ++E+
Sbjct: 2736 VVLLEIVSGREPLNIKRPRTEWSLVEWATPYIRGSKVDEIVDPGIKGGYHAEAMWRVVEV 2915
Query: 562 AARCTDANADDRPSMNQVLQLLEQEVM 588
A +C + + RPSM +++ LE ++
Sbjct: 2916 ALQCLEPFSTYRPSMVAIVRELEDALI 2996
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 203 bits (516), Expect = 7e-53
Identities = 116/302 (38%), Positives = 167/302 (54%), Gaps = 12/302 (3%)
Frame = +1
Query: 307 EIIEKLESLEEEDIVGSGGFGTVYRMVMNDCGTFAVKRIDRSREGCDQVFERELEILGSI 366
E+ + + +E +++G GGFG VY+ + AVK++ Q F E+ +L +
Sbjct: 427 ELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQEFVMEVLMLSLL 606
Query: 367 KHINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHE--NTEQPLNWNDRLNIALGSARGL 424
H NLV L GYC RLL+Y+Y+ +GSL+D L E + ++PLNW+ R+ +A+G+ARGL
Sbjct: 607 HHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAARGL 786
Query: 425 AYLHHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKL-LVDEDAHVTTVVAGTFGYLA 483
YLH P +++RD+KS+NILL+ P +SDFGLAKL V ++ HV+T V GT+GY A
Sbjct: 787 EYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCA 966
Query: 484 PEYLQSGRATEKSDVYSFGVLLLELVTGKRPTDPSFANRGLNVVGWMNTLQKENR----- 538
PEY SG+ T KSD+YSFGV+LLEL+TG+R D S N+V W + R
Sbjct: 967 PEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRRRFGHM 1146
Query: 539 ----LEDVVDRRCTDADAGTLEVILELAARCTDANADDRPSMNQVLQLLEQEVMSPCPSD 594
L+ RC L + + A C RP + ++ LE P
Sbjct: 1147VDPLLQGRFPSRC-------LHQAIAITAMCLQEQPKFRPLITDIVVALEYLASQGSPEA 1305
Query: 595 FY 596
Y
Sbjct: 1306HY 1311
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 201 bits (510), Expect = 4e-52
Identities = 101/216 (46%), Positives = 139/216 (63%), Gaps = 2/216 (0%)
Frame = +2
Query: 303 YTSSEIIEKLESLEEEDIVGSGGFGTVYRMVMNDCGTFAVKRIDRSREGCDQVFERELEI 362
+T E+ +++ +G GGFG+VY +D AVK++ + F E+E+
Sbjct: 266 FTYKELHAATGGFSDDNKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEVEV 445
Query: 363 LGSIKHINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHEN--TEQPLNWNDRLNIALGS 420
LG ++H NL+ LRGYC RL++YDY+ SL LH E LNW R+ IA+GS
Sbjct: 446 LGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQFAVEVQLNWQKRMKIAIGS 625
Query: 421 ARGLAYLHHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLVDEDAHVTTVVAGTFG 480
A G+ YLHHE P I+HRDIK+SN+LLN + EP ++DFG AKL+ + +H+TT V GT G
Sbjct: 626 AEGILYLHHEVTPHIIHRDIKASNVLLNSDFEPLVADFGFAKLIPEGVSHMTTRVKGTLG 805
Query: 481 YLAPEYLQSGRATEKSDVYSFGVLLLELVTGKRPTD 516
YLAPEY G+ +E DVYSFG+LLLELVTG++P +
Sbjct: 806 YLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIE 913
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 199 bits (506), Expect = 1e-51
Identities = 104/246 (42%), Positives = 149/246 (60%), Gaps = 2/246 (0%)
Frame = +1
Query: 303 YTSSEIIEKLESLEEEDIVGSGGFGTVYRMVMNDCGTFAVKRIDRSREGCDQVFERELEI 362
++ E+ + ++ +G GGFG+VY + D AVKR+ D F E+EI
Sbjct: 28 FSLKELHSATNNFNYDNKLGEGGFGSVYWGQLWDGSQIAVKRLKVWSNKADMEFAVEVEI 207
Query: 363 LGSIKHINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLH--ENTEQPLNWNDRLNIALGS 420
L ++H NL++LRGYC RL++YDY+ SL LH ++E L+WN R+NIA+GS
Sbjct: 208 LARVRHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHGQHSSECLLDWNRRMNIAIGS 387
Query: 421 ARGLAYLHHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLVDEDAHVTTVVAGTFG 480
A G+ YLHH+ P I+HRDIK+SN+LL+ + + ++DFG AKL+ D HVTT V GT G
Sbjct: 388 AEGIVYLHHQATPHIIHRDIKASNVLLDSDFQARVADFGFAKLIPDGATHVTTRVKGTLG 567
Query: 481 YLAPEYLQSGRATEKSDVYSFGVLLLELVTGKRPTDPSFANRGLNVVGWMNTLQKENRLE 540
YLAPEY G+A E DV+SFG+LLLEL +GK+P + + ++ W L +
Sbjct: 568 YLAPEYAMLGKANECCDVFSFGILLLELASGKKPLEKLSSTVKRSINDWALPLACAKKFT 747
Query: 541 DVVDRR 546
+ D R
Sbjct: 748 EFADPR 765
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 191 bits (484), Expect = 4e-49
Identities = 109/294 (37%), Positives = 174/294 (59%), Gaps = 10/294 (3%)
Frame = +2
Query: 307 EIIEKLESLEEEDIVGSGGFGTVYRMVMNDCGTFAVKRIDRSREG-CDQVFERELEILGS 365
E+ EK ++ + ++G G +G VY +ND AVK++D S E + F ++ ++
Sbjct: 335 ELKEKTDNFGSKALIGEGSYGRVYYATLNDGNAVAVKKLDVSSEPETNNEFLTQVSMVSR 514
Query: 366 IKHINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHENT----EQP---LNWNDRLNIAL 418
+K+ N V L GYC + R+L Y++ +GSL D+LH QP L+W R+ IA+
Sbjct: 515 LKNDNFVELHGYCVEGNLRVLAYEFATMGSLHDILHGRKGVQGAQPGPTLDWIQRVRIAV 694
Query: 419 GSARGLAYLHHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLVDEDAHV-TTVVAG 477
+ARGL YLH + P I+HRDI+SSN+L+ E+ + I+DF L+ D A + +T V G
Sbjct: 695 DAARGLEYLHEKVQPAIIHRDIRSSNVLIFEDYKAKIADFNLSNQAPDMAARLHSTRVLG 874
Query: 478 TFGYLAPEYLQSGRATEKSDVYSFGVLLLELVTGKRPTDPSFANRGLNVVGWMNTLQKEN 537
TFGY APEY +G+ T+KSDVYSFGV+LLEL+TG++P D + ++V W E+
Sbjct: 875 TFGYHAPEYAMTGQLTQKSDVYSFGVVLLELLTGRKPVDHTMPRGQQSLVTWATPRLSED 1054
Query: 538 RLEDVVDRRCT-DADAGTLEVILELAARCTDANADDRPSMNQVLQLLEQEVMSP 590
+++ VD + + + + +AA C A+ RP+M+ V++ L+ + +P
Sbjct: 1055KVKQCVDPKLKGEYPPKGVAKLAAVAALCVQYEAEFRPNMSIVVKALQPLLKTP 1216
>AI967315
Length = 1308
Score = 189 bits (479), Expect = 1e-48
Identities = 107/290 (36%), Positives = 169/290 (57%), Gaps = 4/290 (1%)
Frame = +1
Query: 303 YTSSEIIEKLESLEEEDIVGSGGFGTVYRMVMNDCGTFAVKRIDRS--REGCDQVFEREL 360
++ E+ E++VG GG+ VY+ + AVKR+ R+ E ++ F E+
Sbjct: 112 FSYEELFHATNGFSSENMVGKGGYAEVYKGRLESGDEIAVKRLTRTCRDERKEKEFLTEI 291
Query: 361 EILGSIKHINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHENTEQPLNWNDRLNIALGS 420
+G + H N++ L G C + + L+++ +GS+ L+H+ PL+W R I LG+
Sbjct: 292 GTIGHVCHSNVMPLLGCC-IDNGLYLVFELSTVGSVASLIHDEKMAPLDWKTRYKIVLGT 468
Query: 421 ARGLAYLHHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLVDEDAHVTTV-VAGTF 479
ARGL YLH C +I+HRDIK+SNILL E+ EP ISDFGLAK L + H + + GTF
Sbjct: 469 ARGLHYLHKGCQRRIIHRDIKASNILLTEDFEPQISDFGLAKWLPSQWTHHSIAPIEGTF 648
Query: 480 GYLAPEYLQSGRATEKSDVYSFGVLLLELVTGKRPTDPSFANRGLNVVGWMNTLQKENRL 539
G+LAPEY G EK+DV++FGV LLE+++G++P D S ++ L+ W + + +
Sbjct: 649 GHLAPEYYMHGVVDEKTDVFAFGVFLLEVISGRKPVDGS--HQSLHT--WAKPILSKWEI 816
Query: 540 EDVVDRRCTDA-DAGTLEVILELAARCTDANADDRPSMNQVLQLLEQEVM 588
E +VD R D + A+ C A++ RP+M++VL+++E+ M
Sbjct: 817 EKLVDPRLEGCYDVTQFNRVAFAASLCIRASSTWRPTMSEVLEVMEEGEM 966
>CN825263
Length = 663
Score = 187 bits (474), Expect = 5e-48
Identities = 101/208 (48%), Positives = 135/208 (64%), Gaps = 3/208 (1%)
Frame = +1
Query: 325 GFGTVYRMVMNDCGTFAVKRIDRSREGCDQVFERELEILGSIKHINLVNLRGYCRLPSAR 384
GFG VY+ ++ND AVK + R + + F E+E+L + H NLV L G C R
Sbjct: 1 GFGLVYKGILNDGRDVAVKILKRDDQRGGREFLAEVEMLSRLHHRNLVKLIGICIEKQTR 180
Query: 385 LLIYDYLAIGSLDDLLHENTEQ--PLNWNDRLNIALGSARGLAYLHHECCPKIVHRDIKS 442
LIY+ + GS++ LH ++ PL+WN R+ IALG+ARGLAYLH + P ++HRD KS
Sbjct: 181 CLIYELVPNGSVESHLHGADKETGPLDWNARMKIALGAARGLAYLHEDSNPCVIHRDFKS 360
Query: 443 SNILLNENMEPHISDFGLAKLLVDE-DAHVTTVVAGTFGYLAPEYLQSGRATEKSDVYSF 501
SNILL + P +SDFGLA+ +DE + H++T V GTFGYLAPEY +G KSDVYS+
Sbjct: 361 SNILLECDFTPKVSDFGLARTALDEGNKHISTHVMGTFGYLAPEYAMTGHLLVKSDVYSY 540
Query: 502 GVLLLELVTGKRPTDPSFANRGLNVVGW 529
GV+LLEL+TG +P D S N+V W
Sbjct: 541 GVVLLELLTGTKPVDLSQPPGQENLVTW 624
>TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like
protein, partial (46%)
Length = 941
Score = 184 bits (468), Expect = 3e-47
Identities = 102/267 (38%), Positives = 157/267 (58%), Gaps = 2/267 (0%)
Frame = +3
Query: 320 IVGSGGFGTVYRMVMNDCGTFAVKRIDRSREGCDQVFERELEILGSIKHINLVNLRGYCR 379
++G GGFG VY ++ A+KR + E F+ E+E+L ++H +LV+L GYC
Sbjct: 12 LLGVGGFGKVYYGEVDGGTKVAIKRGNPLSEQGVHEFQTEIEMLSKLRHRHLVSLIGYCE 191
Query: 380 LPSARLLIYDYLAIGSLDDLLHENTEQPLNWNDRLNIALGSARGLAYLHHECCPKIVHRD 439
+ +L+YD++A G+L + L++ + PL W RL I +G+ARGL YLH I+HRD
Sbjct: 192 ENTEMILVYDHMAYGTLREHLYKTQKPPLPWKQRLEICIGAARGLHYLHTGAKYTIIHRD 371
Query: 440 IKSSNILLNENMEPHISDFGLAKL-LVDEDAHVTTVVAGTFGYLAPEYLQSGRATEKSDV 498
+K++NILL+E +SDFGL+K ++ HV+TVV G+FGYL PEY + + T+KSDV
Sbjct: 372 VKTTNILLDEKWVAKVSDFGLSKTGPTLDNTHVSTVVKGSFGYLDPEYFRRQQLTDKSDV 551
Query: 499 YSFGVLLLELVTGKRPTDPSFANRGLNVVGWMNTLQKENRLEDVVDRRCTDADA-GTLEV 557
YSFGV+L E++ + +PS A +++ W + + L+ ++D A +
Sbjct: 552 YSFGVVLFEILCARPALNPSLAKEQVSLAEWASHCYNKGILDQILDPYLKGKIAPECFKK 731
Query: 558 ILELAARCTDANADDRPSMNQVLQLLE 584
E A +C +RPSM VL LE
Sbjct: 732 FAETAMKCVSDQGIERPSMGDVLWNLE 812
>TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like protein kinase
protein (AT3g17840/MEB5_6), partial (45%)
Length = 1489
Score = 171 bits (433), Expect = 3e-43
Identities = 106/281 (37%), Positives = 156/281 (54%), Gaps = 5/281 (1%)
Frame = +3
Query: 319 DIVGSGGFGTVYRMVMNDCGTFAVKRIDRSREGCDQVFERELEILGSIKHINLVNLRGYC 378
+++G G FGT Y+ V+ AVKR+ + ++ F+ ++E +G++ H +LV LR Y
Sbjct: 288 EVLGKGTFGTAYKAVLETGLVVAVKRL-KDVTISEKEFKDKIETVGAMDHQSLVPLRAYY 464
Query: 379 RLPSARLLIYDYLAIGSLDDLLHENT---EQPLNWNDRLNIALGSARGLAYLHHECCPKI 435
+LL+YDY+ +GSL LLH N PLNW R IALG+ARG+ YLH + P +
Sbjct: 465 FSRDEKLLVYDYMPMGSLSALLHGNKGAGRTPLNWEIRSGIALGAARGIEYLHSQG-PNV 641
Query: 436 VHRDIKSSNILLNENMEPHISDFGLAKLLVDEDAHVTTVVAGTFGYLAPEYLQSGRATEK 495
H +IK+SNILL ++ E +SDFGLA L+ GY APE + ++K
Sbjct: 642 SHGNIKASNILLTKSYEAKVSDFGLAHLVGPSSTPNRVA-----GYRAPEVTDPRKVSQK 806
Query: 496 SDVYSFGVLLLELVTGKRPTDPSFANRGLNVVGWMNTLQKENRLEDVVDRRCTDADAGTL 555
+DVYSFGVLLLEL+TGK PT G+++ W+ +L +E +V D
Sbjct: 807 ADVYSFGVLLLELLTGKAPTHALLNEEGVDLPRWVQSLVREEWTSEVFDLELLRYQNVEE 986
Query: 556 EVI--LELAARCTDANADDRPSMNQVLQLLEQEVMSPCPSD 594
E++ L+LA C D RPS+ +V + +E+ S D
Sbjct: 987 EMVQLLQLAVDCAAPYPDRRPSIAEVTRSIEELCRSNLKED 1109
>TC16248 similar to UP|BAC87845 (BAC87845) Leucine-rich repeat receptor-like
protein kinase 1, partial (17%)
Length = 866
Score = 163 bits (412), Expect = 8e-41
Identities = 91/229 (39%), Positives = 130/229 (56%), Gaps = 4/229 (1%)
Frame = +2
Query: 359 ELEILGSIKHINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHENTEQP-LNWNDRLNIA 417
E++ L I+H N++ L G+C L+Y++L GSLD +L+ +T+ +W R+N+
Sbjct: 5 EIQTLTEIRHRNVIKLHGFCLHSKFSFLVYEFLEGGSLDQVLNSDTQAAAFHWEKRVNVV 184
Query: 418 LGSARGLAYLHHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLVDEDAHVTTVVAG 477
G A L+Y+HH+C P I+HRDI S N+LL+ E H+SDFG AK L +H T AG
Sbjct: 185 KGVANALSYMHHDCSPPIIHRDISSKNVLLDLQYEAHVSDFGTAKFL-KPGSHTWTAFAG 361
Query: 478 TFGYLAPEYLQSGRATEKSDVYSFGVLLLELVTGKRPTDPSFANRGLNVVGWMNTLQKEN 537
TFGY APE Q+ + EK DVYSFGV LE++ G P D + + V +N L
Sbjct: 362 TFGYAAPELAQTMQVNEKCDVYSFGVFALEIIMGMHPGDLISSLMSPSTVPMVNDL---- 529
Query: 538 RLEDVVDRRCTDADAGTLE---VILELAARCTDANADDRPSMNQVLQLL 583
L D++D+R + E +I LA C N RP+M+QV + L
Sbjct: 530 LLIDILDQRPHQVEKPIDEEVILIASLALACLRKNPHSRPTMDQVSKAL 676
>TC12493 similar to UP|Q9LP77 (Q9LP77) T1N15.9, partial (35%)
Length = 837
Score = 163 bits (412), Expect = 8e-41
Identities = 94/230 (40%), Positives = 137/230 (58%), Gaps = 8/230 (3%)
Frame = +3
Query: 309 IEKLESLEE-----EDIVGSGGFGTVYRMVMNDCGTFAVKRIDRSREGCDQVFERELEIL 363
++++ SL+E +++G G FGT Y+ M + AVKR+ + + F ++E +
Sbjct: 165 VDRVFSLDELLRASAEVLGKGTFGTTYKATMEMGRSVAVKRL-KDVTATEMEFREKIEEV 341
Query: 364 GSIKHINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHENT---EQPLNWNDRLNIALGS 420
G + H NLV LRGY +L++YDY+ +GSL LLH N PLNW R IALG+
Sbjct: 342 GKLVHENLVPLRGYYFSRDEKLVVYDYMPMGSLSALLHANNGAGRTPLNWETRSAIALGA 521
Query: 421 ARGLAYLHHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLVDEDAHVTTVVAGTFG 480
A G+AYLH + P H +IKSSNILL ++ EP +SD GLA L + T+ G
Sbjct: 522 AHGIAYLHSQG-PTSSHGNIKSSNILLTKSFEPRVSDLGLAYLALP-----TSTPNRISG 683
Query: 481 YLAPEYLQSGRATEKSDVYSFGVLLLELVTGKRPTDPSFANRGLNVVGWM 530
Y APE + + ++K+DVYSFG++LLEL+TGK PT S G+++ W+
Sbjct: 684 YRAPEVTDARKVSQKADVYSFGIMLLELLTGKPPTHSSLNEEGVDLPRWV 833
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 163 bits (412), Expect = 8e-41
Identities = 119/393 (30%), Positives = 195/393 (49%), Gaps = 31/393 (7%)
Frame = +1
Query: 222 IPHAESDEAAVPTKRSSSHYMKVVLIGAMTTLGLLLLVTLSF-LWIRLLSKKERAVMRYT 280
IP + VP ++ +G ++ G +L+ L+F +++R K+E T
Sbjct: 739 IPGRYKNGVYVPLYHRTAGLASGAAVG-ISIAGTFVLLLLAFCMYVRYQKKEEEKAKLPT 915
Query: 281 DVKKQVDPE-----ASTKLITFHGDLPYTSS------------------EIIEKLESLEE 317
D+ + + +S + T P T+S E+ + +
Sbjct: 916 DISMALSTQDGNASSSAEYETSGSSGPGTASATGLTSIMVAKSMEFSYQELAKATNNFSL 1095
Query: 318 EDIVGSGGFGTVYRMVMNDCGTFAVKRIDRSREGCDQVFERELEILGSIKHINLVNLRGY 377
++ +G GGFG VY + T A+K++D F EL++L + H+NLV L GY
Sbjct: 1096 DNKIGQGGFGAVYYAELRGKKT-AIKKMDVQ---ASTEFLCELKVLTHVHHLNLVRLIGY 1263
Query: 378 CRLPSARLLIYDYLAIGSLDDLLHENTEQPLNWNDRLNIALGSARGLAYLHHECCPKIVH 437
C + + L+Y+++ G+L LH + ++PL W+ R+ IAL +ARGL Y+H P +H
Sbjct: 1264 C-VEGSLFLVYEHIDNGNLGQYLHGSGKEPLPWSSRVQIALDAARGLEYIHEHTVPVYIH 1440
Query: 438 RDIKSSNILLNENMEPHISDFGLAKLLVDEDAHVTTVVAGTFGYLAPEYLQSGRATEKSD 497
RD+KS+NIL+++N+ ++DFGL KL+ ++ + T + GTFGY+ PEY Q G + K D
Sbjct: 1441 RDVKSANILIDKNLRGKVADFGLTKLIEVGNSTLQTRLVGTFGYMPPEYAQYGDISPKID 1620
Query: 498 VYSFGVLLLELVTGKR---PTDPSFANRGLNVVGWMNTLQKE---NRLEDVVDRRC-TDA 550
VY+FGV+L EL++ K T A V + L K + L +VD R +
Sbjct: 1621 VYAFGVVLFELISAKNAVLKTGELVAESKGLVALFEEALNKSDPCDALRKLVDPRLGENY 1800
Query: 551 DAGTLEVILELAARCTDANADDRPSMNQVLQLL 583
++ I +L CT N RPSM ++ L
Sbjct: 1801 PIDSVLKIAQLGRACTRDNPLLRPSMRSLVVAL 1899
>TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P14.15
{Arabidopsis thaliana;}, partial (37%)
Length = 1079
Score = 157 bits (396), Expect = 6e-39
Identities = 90/224 (40%), Positives = 135/224 (60%), Gaps = 3/224 (1%)
Frame = +3
Query: 359 ELEILGSIKHINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHENTEQPLNWNDRLNIAL 418
E+E+L I H NLV L G+ + R+LI +Y+ G+L + L + L++N RL IA+
Sbjct: 6 EVELLAKIDHRNLVKLLGFIDKGNERILITEYVPNGTLREHLDGLRGKILDFNQRLEIAI 185
Query: 419 GSARGLAYLHHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKL--LVDEDAHVTTVVA 476
A GL YLH +I+HRD+KSSNILL E+M ++DFG A+L + + H++T V
Sbjct: 186 DVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFARLGPVNGDQTHISTKVK 365
Query: 477 GTFGYLAPEYLQSGRATEKSDVYSFGVLLLELVTGKRPTDPSFANRGLNVVGWMNTLQKE 536
GT GYL PEY+++ + T KSDVYSFG+LLLE++TG+RP + A + W E
Sbjct: 366 GTVGYLDPEYMKTHQLTPKSDVYSFGILLLEILTGRRPVELKKAADERVTLRWAFRKYNE 545
Query: 537 NRLEDVVDRRCTDA-DAGTLEVILELAARCTDANADDRPSMNQV 579
+ +++D +A +A L +L+L+ +C DRP+M V
Sbjct: 546 GSVVELMDPLMEEAVNADVLMKMLDLSFQCAAPIRTDRPNMKSV 677
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 154 bits (388), Expect = 5e-38
Identities = 99/286 (34%), Positives = 157/286 (54%), Gaps = 5/286 (1%)
Frame = +1
Query: 321 VGSGGFGTVYRMVMNDCGTFAVKRIDRSREGCDQVFERELEILGSIKHINLVNLRGYCRL 380
+G GGFG VY+ ++ + AVKR+ + + F+ E++++ ++H NLV L G C +
Sbjct: 1690 LGEGGFGPVYKGLLANGQEIAVKRLSNTSGQGMEEFKNEIKLIARLQHRNLVKLFG-CSV 1866
Query: 381 PSARLLIYDYLAIGSLDDLLHENTEQPLNWNDRLNIALGSARGLAYLHHECCPKIVHRDI 440
+ A + LL + ++WN RL I G ARGL YLH + +I+HRD+
Sbjct: 1867 HQDE----NSHANKKMKILLDSTRSKLVDWNKRLQIIDGIARGLLYLHQDSRLRIIHRDL 2034
Query: 441 KSSNILLNENMEPHISDFGLAKLLVDEDAHVTTV-VAGTFGYLAPEYLQSGRATEKSDVY 499
K+SNILL++ M P ISDFGLA++ + + T V GT+GY+ PEY G + KSDV+
Sbjct: 2035 KTSNILLDDEMNPKISDFGLARIFIGDQVEARTKRVMGTYGYMPPEYAVHGSFSIKSDVF 2214
Query: 500 SFGVLLLELVTGKRPTDPSFANRGLNVVGWMNTLQKENRLEDVVDRRCTDADAGTLEVI- 558
SFGV++LE+++GK+ + LN++ L E R ++VD D T E++
Sbjct: 2215 SFGVIVLEIISGKKIGRFYDPHHHLNLLSHAWRLWIEERPLELVDELLDDPVIPT-EILR 2391
Query: 559 -LELAARCTDANADDRPSMNQVLQLL--EQEVMSPCPSDFYESHSD 601
+ +A C ++RP M ++ +L E+E+ P FY D
Sbjct: 2392 YIHVALLCVQRRPENRPDMLSIVLMLNGEKELPKPRLPAFYTGKHD 2529
>TC17311 similar to UP|Q8K2Y2 (Q8K2Y2) Receptor-interacting serine-threonine
kinase 3, partial (7%)
Length = 606
Score = 152 bits (383), Expect = 2e-37
Identities = 80/165 (48%), Positives = 108/165 (64%), Gaps = 11/165 (6%)
Frame = +3
Query: 359 ELEILGSIKHINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHENTEQP----------L 408
E+EIL +I+H N+V L+ + LL+YDYL SLD LH+ ++ P +
Sbjct: 3 EVEILSNIRHNNIVKLQCCISSEESLLLVYDYLENLSLDRWLHKKSKPPTVSGSVQNNII 182
Query: 409 NWNDRLNIALGSARGLAYLHHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLV-DE 467
+W RL+IA+G A+GL Y+HH+C P +VHRD+K SNILL+ ++DFGLA + V E
Sbjct: 183 DWPRRLHIAIGVAQGLCYMHHDCSPPVVHRDLKPSNILLDSQFNAKVADFGLAMMSVKPE 362
Query: 468 DAHVTTVVAGTFGYLAPEYLQSGRATEKSDVYSFGVLLLELVTGK 512
+ + VAGTFGY+APEY + R EK DVYSFGV+LLEL TGK
Sbjct: 363 ELATMSAVAGTFGYIAPEYALTIRVNEKIDVYSFGVILLELTTGK 497
>TC9538 similar to UP|RLK5_ARATH (P47735) Receptor-like protein kinase 5
precursor , partial (10%)
Length = 1067
Score = 152 bits (383), Expect = 2e-37
Identities = 88/237 (37%), Positives = 134/237 (56%), Gaps = 23/237 (9%)
Frame = +2
Query: 370 NLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHENTEQP-----------LNWNDRLNIAL 418
N+V L ++ LL+Y+YL SLD LH + L+W RL IA+
Sbjct: 5 NIVRLLCCISNEASMLLVYEYLENHSLDKWLHLKPKSSSVSGVVQQYTVLDWPKRLKIAI 184
Query: 419 GSARGLAYLHHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLVDE-DAHVTTVVAG 477
G+A+GL+Y+HH+C P IVHRD+K+SNILL++ ++DFGLA++L+ + ++ + V G
Sbjct: 185 GAAQGLSYMHHDCSPPIVHRDVKTSNILLDKQFNAKVADFGLARMLIKPGELNIMSTVIG 364
Query: 478 TFGYLAPEYLQSGRATEKSDVYSFGVLLLELVTGKRPTDPSFANRGLNVVGW-------- 529
TFGY+APEY+Q+ R +EK DVYSFGV+LLEL TGK + ++ ++ ++ W
Sbjct: 365 TFGYIAPEYVQTTRISEKVDVYSFGVVLLELTTGK---EANYGDQHSSLAEWAWRHILIG 535
Query: 530 ---MNTLQKENRLEDVVDRRCTDADAGTLEVILELAARCTDANADDRPSMNQVLQLL 583
+ L K+ +D C+ + +L CT RPSM +VL +L
Sbjct: 536 SNVXDLLXKDVMEASYIDEMCS---------VFKLGVMCTATLPATRPSMKEVLPIL 679
>TC15999 similar to UP|Q9LLT5 (Q9LLT5) Receptor-like protein kinase
(Fragment), complete
Length = 999
Score = 149 bits (377), Expect = 1e-36
Identities = 94/258 (36%), Positives = 140/258 (53%), Gaps = 36/258 (13%)
Frame = +1
Query: 363 LGSIKHINLVNLRGYCRLPSARLLIYDYLAIGSLDDLLHE---------------NTEQP 407
LG IKH NLV L GYC R+ IYDY+ G+L +LL++ E+P
Sbjct: 4 LGRIKHPNLVLLTGYCLAGDQRIAIYDYMENGNLQNLLYDLPLGVLHSTDDWSTDTWEEP 183
Query: 408 LN--------------WNDRLNIALGSARGLAYLHHECCPKIVHRDIKSSNILLNENMEP 453
N W+ R IALG+AR LA+LHH C P I+HR +K+S++ L+ ++EP
Sbjct: 184 DNNGIQNAGSEGLLTTWSFRHKIALGTARALAFLHHGCSPPIIHRAVKASSVYLDYDLEP 363
Query: 454 HISDFGLAKLL---VDEDAHVTTVVAGTFGYLAPEYLQS--GRATEKSDVYSFGVLLLEL 508
+SDFGLAK+ +DE+ + G+ GY PE+ Q T KSDVY FGV+L EL
Sbjct: 364 RLSDFGLAKIFGSGLDEE-----IARGSPGYDPPEFTQPDFDTPTTKSDVYCFGVVLFEL 528
Query: 509 VTGKRPTDPSF-ANRGLNVVGWMNTLQKENRLEDVVDRRCTDADAG-TLEVILELAARCT 566
+TGK+P + + ++ +V W+ L ++N+ +D + D +E L++ CT
Sbjct: 529 LTGKKPVEDDYHDDKEETLVSWVRGLVRKNQTSRAIDPKIRDTGPDEQMEEALKIGYLCT 708
Query: 567 DANADDRPSMNQVLQLLE 584
RP+M Q++ LL+
Sbjct: 709 ADLPFKRPTMQQIVGLLK 762
>CB827497
Length = 550
Score = 146 bits (368), Expect = 1e-35
Identities = 84/175 (48%), Positives = 112/175 (64%), Gaps = 2/175 (1%)
Frame = +2
Query: 406 QPLNWNDRLNIALGSARGLAYLHHECCPKIVHRDIKSSNILLNENMEPHISDFGLAKLLV 465
+ L W R NIALG+ARGL YLH C +I+HRDIK++NILL E+ EP I DFGLAK L
Sbjct: 2 EKLPWCIRHNIALGTARGLLYLHEGCQRRIIHRDIKAANILLTEDYEPQICDFGLAKWLP 181
Query: 466 DEDAHVTTV-VAGTFGYLAPEYLQSGRATEKSDVYSFGVLLLELVTGKRPTDPSFANRGL 524
+ H T GTFGYLAPEYL G EK+DV++FGVLLLELV+G+R D F+ + L
Sbjct: 182 ENWTHHTVAKFEGTFGYLAPEYLLHGIVDEKTDVFAFGVLLLELVSGRRALD--FSQQSL 355
Query: 525 NVVGWMNTLQKENRLEDVVDRRCTDA-DAGTLEVILELAARCTDANADDRPSMNQ 578
V W L K+N + +++D D D+ + ++L A+ C ++ RPS+ Q
Sbjct: 356 --VLWAKPLLKKNEIMELIDPSLADGFDSRQMNLMLLAASLCIQQSSIRRPSIRQ 514
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.138 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,352,321
Number of Sequences: 28460
Number of extensions: 135981
Number of successful extensions: 1311
Number of sequences better than 10.0: 376
Number of HSP's better than 10.0 without gapping: 1009
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1082
length of query: 603
length of database: 4,897,600
effective HSP length: 96
effective length of query: 507
effective length of database: 2,165,440
effective search space: 1097878080
effective search space used: 1097878080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0170.5


