

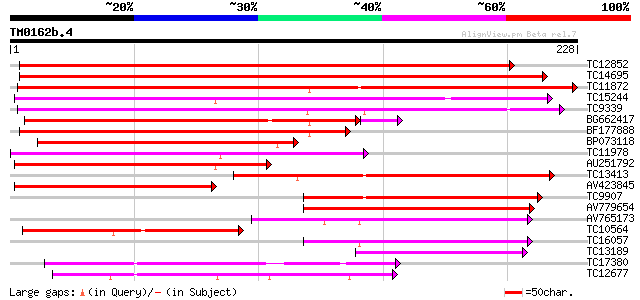
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0162b.4
(228 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC12852 similar to UP|Q9FQD9 (Q9FQD9) Glutathione S-transferase ... 227 1e-60
TC14695 similar to UP|Q9FQE5 (Q9FQE5) Glutathione S-transferase ... 226 3e-60
TC11872 similar to UP|GTX6_SOYBN (P32110) Probable glutathione S... 217 1e-57
TC15244 similar to UP|O49235 (O49235) 2,4-D inducible glutathion... 173 2e-44
TC9339 similar to UP|Q9FQE3 (Q9FQE3) Glutathione S-transferase G... 169 5e-43
BG662417 141 8e-35
BF177888 137 2e-33
BP073118 130 2e-31
TC11978 similar to UP|GTXA_TOBAC (P25317) Probable glutathione S... 122 5e-29
AU251792 118 7e-28
TC13413 similar to UP|GTX6_SOYBN (P32110) Probable glutathione S... 106 3e-24
AV423845 99 5e-22
TC9907 weakly similar to UP|Q84T17 (Q84T17) Glutathione S-transf... 90 3e-19
AV779654 77 2e-15
AV765173 74 2e-14
TC10564 similar to UP|Q9FQD3 (Q9FQD3) Glutathione S-transferase ... 67 3e-12
TC16057 weakly similar to UP|Q9M6R4 (Q9M6R4) Glutathione S-trans... 63 4e-11
TC13189 similar to UP|Q9FUS9 (Q9FUS9) Glutathione S-transferase,... 56 5e-09
TC17380 similar to UP|Q84UH4 (Q84UH4) Dehydroascorbate reductase... 45 1e-05
TC12677 similar to UP|Q9FQD6 (Q9FQD6) Glutathione S-transferase ... 42 1e-04
>TC12852 similar to UP|Q9FQD9 (Q9FQD9) Glutathione S-transferase GST 19 ,
partial (97%)
Length = 909
Score = 227 bits (579), Expect = 1e-60
Identities = 103/199 (51%), Positives = 147/199 (73%)
Frame = +2
Query: 5 DVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHKT 64
+VKL++ S R+EWAL++KGVEYEY++ED+ NKS LLL+ NPVHKK+PVL+H K
Sbjct: 65 EVKLIATPQSFPCARIEWALRIKGVEYEYIQEDLANKSPLLLQSNPVHKKVPVLLHDGKP 244
Query: 65 IAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQEKA 124
IAES +I+EYIDETWK+ PLLP DPYERALARFWA F ++K + V+ A G+E+ KA
Sbjct: 245 IAESLVILEYIDETWKENPLLPLDPYERALARFWARFIDEKCVIGVWGACVAQGEEKGKA 424
Query: 125 LNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKFPAT 184
+ A +++E +E+ I+G ++FGGE IGYLDI GW+S+WI++ EE+G + ++ +FP+
Sbjct: 425 IETALDSLEFLEKQIQGNKYFGGERIGYLDIVAGWLSHWINVLEELGDMELLTAERFPSL 604
Query: 185 TAWMTNFLSHPVIKDTLPP 203
W NF+ ++D +PP
Sbjct: 605 HEWSQNFIQTSPVQDCIPP 661
>TC14695 similar to UP|Q9FQE5 (Q9FQE5) Glutathione S-transferase GST 13 ,
partial (97%)
Length = 1338
Score = 226 bits (575), Expect = 3e-60
Identities = 105/212 (49%), Positives = 146/212 (68%)
Frame = +3
Query: 5 DVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHKT 64
+V+LL W SPFS RV+ ALKLKGV Y Y EED+ NKS+ LL+ NPV+KK+PVLVH
Sbjct: 93 EVRLLGKWASPFSNRVDLALKLKGVPYNYSEEDLANKSADLLKYNPVYKKVPVLVHNGNP 272
Query: 65 IAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQEKA 124
+AES +I+EYIDETWK P LP DPYERA+ARFW+ + K+L ++ A +EKA
Sbjct: 273 LAESLVILEYIDETWKNSPFLPQDPYERAVARFWSKTLDDKILPPIWNACWSDEKGREKA 452
Query: 125 LNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKFPAT 184
+ EA EA++ +++ ++ RFFGGE IG +DIA +I YW+ I +E+ + ++ KFP
Sbjct: 453 VEEALEALKLLQDQLKDNRFFGGETIGLVDIAANFIGYWVSILQEIAGLELLTVEKFPKL 632
Query: 185 TAWMTNFLSHPVIKDTLPPRDKMIDYFHGRKK 216
W F++HPVIK+ LPPRD++ +F +K
Sbjct: 633 FKWSQEFVNHPVIKEGLPPRDELFAFFKASQK 728
>TC11872 similar to UP|GTX6_SOYBN (P32110) Probable glutathione
S-transferase (Heat shock protein 26A) (G2-4) ,
complete
Length = 968
Score = 217 bits (553), Expect = 1e-57
Identities = 105/226 (46%), Positives = 159/226 (69%), Gaps = 1/226 (0%)
Frame = +2
Query: 4 QDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHK 63
++V+LL SPF RV+ AL LKGVEYE+VE+ KS LLL+ NPVHKK+PV VH K
Sbjct: 38 ENVQLLGIVGSPFVCRVQIALNLKGVEYEFVEQIFGQKSDLLLKYNPVHKKVPVFVHNDK 217
Query: 64 TIAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGD-EQE 122
IAES +I+EYIDETWK P+LP DPY+RALARFW+ F + K+++T + A+ I + E+E
Sbjct: 218 PIAESLVILEYIDETWKSNPILPSDPYKRALARFWSKFIDDKVIDTTWKAVFILEEAERE 397
Query: 123 KALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKFP 182
K + E EA++ +E ++ ++FFGGE ++DIA +I+++I +++EV +H++ FP
Sbjct: 398 KIIEELSEALQYLENELK-EKFFGGEEFEFVDIAAVFIAFYIPLFQEVAGLHLLTAENFP 574
Query: 183 ATTAWMTNFLSHPVIKDTLPPRDKMIDYFHGRKKDLSETFRGRFKV 228
W +FL+HPV+K+++PPRD ++ F GR + L+ + +F+V
Sbjct: 575 KLYNWSQDFLNHPVVKESMPPRDPLLAIFKGRYESLTASK*DQFRV 712
>TC15244 similar to UP|O49235 (O49235) 2,4-D inducible glutathione
S-transferase , complete
Length = 1309
Score = 173 bits (439), Expect = 2e-44
Identities = 92/217 (42%), Positives = 131/217 (59%), Gaps = 1/217 (0%)
Frame = +3
Query: 3 SQDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGH 62
+ +V LL FW SPF RV AL KG+ YEY EED+ NKS LLL+ NPVHKKIPVL+H
Sbjct: 48 ADEVVLLDFWASPFGMRVRIALAEKGINYEYKEEDLRNKSPLLLQSNPVHKKIPVLIHNG 227
Query: 63 KTIAESFIIIEYIDETWKQ-YPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQ 121
K IAES I ++YIDE W PLLP DPY+RA ARFWA++ ++K+ E + +E+
Sbjct: 228 KPIAESLIAVQYIDEVWNDASPLLPSDPYQRAQARFWADYVDKKIYEIGRNIIWKKNEER 407
Query: 122 EKALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKF 181
E A E + ++ +EE + K +FGG+ +GY+DIA+ W +E +G+ +I +
Sbjct: 408 EAAKKEFIDCLKLLEEQLGDKTYFGGDKLGYVDIALVPFYTWFKGYETLGNFNI--ESEC 581
Query: 182 PATTAWMTNFLSHPVIKDTLPPRDKMIDYFHGRKKDL 218
P AW L + +L +DK+ ++ +K L
Sbjct: 582 PKLIAWAKRCLQKESVSKSLHDQDKIYEFIVEIRKKL 692
>TC9339 similar to UP|Q9FQE3 (Q9FQE3) Glutathione S-transferase GST 15
(Fragment) , partial (44%)
Length = 981
Score = 169 bits (427), Expect = 5e-43
Identities = 90/223 (40%), Positives = 130/223 (57%), Gaps = 3/223 (1%)
Frame = +3
Query: 4 QDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHK 63
+ +++ FW SPF+ RV W LKLKG+ +EY EED +NKS LL+ NPVHKK PVLVH K
Sbjct: 12 EQIEVHGFWYSPFTFRVLWTLKLKGIAFEYFEEDRYNKSPQLLQYNPVHKKTPVLVHDGK 191
Query: 64 TIAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITG-DEQE 122
+ ES II+EYIDE W Q PL+P DPYE+A+ARFW + + + + G +EQ+
Sbjct: 192 PLCESMIIVEYIDELWPQNPLVPSDPYEKAVARFWVRYVDDMMSPVLPQFFGRFGSEEQD 371
Query: 123 KALNEAREAMEKIEEVIEG--KRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLK 180
K + + E E IE+ G K+F GG+ + +DI G + ++ + I+ K
Sbjct: 372 KVIKDIWERFEVIEDQYLGDHKKFLGGDTLNIVDITFGSFMKTLVGMQDALEVKILVAEK 551
Query: 181 FPATTAWMTNFLSHPVIKDTLPPRDKMIDYFHGRKKDLSETFR 223
FP AW NF+ PVI + P +K++ ++ LS + R
Sbjct: 552 FPRLHAWFNNFMDVPVINNN-PEHEKLVAAMKVFREKLSASSR 677
>BG662417
Length = 484
Score = 141 bits (355), Expect(2) = 8e-35
Identities = 69/137 (50%), Positives = 89/137 (64%), Gaps = 2/137 (1%)
Frame = +2
Query: 7 KLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHKTIA 66
KL FW SPF RV W LK+KG+ YEY+EED NK+ LLE +PVHKK+PV VH K +
Sbjct: 17 KLHGFWFSPFVLRVVWTLKVKGIPYEYIEEDPANKTPQLLEYHPVHKKVPVFVHDGKPVC 196
Query: 67 ESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGD--EQEKA 124
ES II+EYIDE W Q PL+P DPYERA+ARFW + + ++ V + +C + D E+E+
Sbjct: 197 ESIIIVEYIDEIWSQNPLVPADPYERAIARFWVRYIDD-MISAVLLPLCRSNDIAEREEI 373
Query: 125 LNEAREAMEKIEEVIEG 141
+ E IEE G
Sbjct: 374 IKEIWARFRVIEEQCSG 424
Score = 21.6 bits (44), Expect(2) = 8e-35
Identities = 8/17 (47%), Positives = 10/17 (58%)
Frame = +1
Query: 142 KRFFGGENIGYLDIAVG 158
K F GG+ +DIA G
Sbjct: 430 KEFLGGDTFNIVDIAFG 480
>BF177888
Length = 420
Score = 137 bits (345), Expect = 2e-33
Identities = 68/134 (50%), Positives = 85/134 (62%), Gaps = 1/134 (0%)
Frame = +2
Query: 5 DVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHKT 64
D+KL FW SPF+ RV W LKLKG+ +EY+EED FNKS LL+ NPVHKK PV VH K
Sbjct: 17 DLKLHGFWYSPFTFRVLWTLKLKGIPFEYIEEDRFNKSPQLLQYNPVHKKTPVFVHDGKP 196
Query: 65 IAESFIIIEYIDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGD-EQEK 123
I ES II+EYIDE W Q PL+P DP+ +A+ARFW + E + G E+EK
Sbjct: 197 ICESMIIVEYIDELWTQNPLVPSDPHVKAVARFWVRYVEDMISAVASPLFRSNGSAEREK 376
Query: 124 ALNEAREAMEKIEE 137
+ + E IE+
Sbjct: 377 VIKDIWERFGVIED 418
>BP073118
Length = 495
Score = 130 bits (326), Expect = 2e-31
Identities = 60/107 (56%), Positives = 77/107 (71%), Gaps = 2/107 (1%)
Frame = +1
Query: 12 WPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHKTIAESFII 71
W SPF+ RV W LKLKG+ +EY+EED +NKS LL+ NPVHKK PVLVH K + ES II
Sbjct: 25 WYSPFTCRVLWTLKLKGIAFEYLEEDRYNKSPQLLQYNPVHKKTPVLVHDGKPLCESMII 204
Query: 72 IEYIDETWKQYPLLPHDPYERALARFWANFTEQKL--LETVFVAMCI 116
+EYIDE W Q PL+P DPYE+A+ARFW + + + L+T + C+
Sbjct: 205 VEYIDELWPQNPLVPSDPYEKAVARFWVRYVDDMVHFLQT*HLYACV 345
>TC11978 similar to UP|GTXA_TOBAC (P25317) Probable glutathione
S-transferase parA (Auxin-regulated protein parA)
(STR246C protein) , partial (65%)
Length = 454
Score = 122 bits (306), Expect = 5e-29
Identities = 64/145 (44%), Positives = 87/145 (59%), Gaps = 1/145 (0%)
Frame = +2
Query: 1 MGSQDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVH 60
M V LL FWPS + RV+ AL KG+ YE +ED KSSLLLE+NPVHK IPVL+H
Sbjct: 20 MAENKVVLLDFWPSSYGMRVKIALAEKGISYECKQEDFQAKSSLLLEMNPVHKMIPVLIH 199
Query: 61 GHKTIAESFIIIEYIDETWKQYP-LLPHDPYERALARFWANFTEQKLLETVFVAMCITGD 119
K I ES I+EYIDE W P LLP DPY+R+ ARFW ++ ++ + G+
Sbjct: 200 NGKPICESLNIVEYIDEAWNHKPSLLPADPYKRSQARFWGDYIDKNVYSIGKRVWTGKGE 379
Query: 120 EQEKALNEAREAMEKIEEVIEGKRF 144
+QE+ + E ++ +E + K +
Sbjct: 380 DQEEGKKKFIECLKILEGELGDKPY 454
>AU251792
Length = 363
Score = 118 bits (296), Expect = 7e-28
Identities = 55/104 (52%), Positives = 70/104 (66%), Gaps = 1/104 (0%)
Frame = +2
Query: 3 SQDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGH 62
S DVKLL W SP + R AL +K V+YE++ E KS LLL+ NPVH+K+PV H
Sbjct: 29 SDDVKLLGAWQSPVTLRARIALNIKSVDYEFIAESFEPKSQLLLQSNPVHQKVPVFFHKD 208
Query: 63 KTIAESFIIIEYIDETWKQ-YPLLPHDPYERALARFWANFTEQK 105
K I ES +I++Y+DE W P+LP DPYERA+ARFWA F + K
Sbjct: 209 KPICESLVIVQYVDEVWSSASPILPSDPYERAIARFWAAFIDDK 340
>TC13413 similar to UP|GTX6_SOYBN (P32110) Probable glutathione
S-transferase (Heat shock protein 26A) (G2-4) , partial
(59%)
Length = 535
Score = 106 bits (265), Expect = 3e-24
Identities = 45/130 (34%), Positives = 88/130 (67%), Gaps = 1/130 (0%)
Frame = +1
Query: 91 ERALARFWANFTEQKLLETVFVAM-CITGDEQEKALNEAREAMEKIEEVIEGKRFFGGEN 149
+RA+ARFW+ + K+++T + ++ + +E+EK + E EA++ +E ++ + FFGGE
Sbjct: 1 QRAMARFWSKVIDDKVVDTTWKSVFTVEEEEREKRIEELSEALQFLENELKDQ-FFGGEE 177
Query: 150 IGYLDIAVGWISYWIHIWEEVGSIHIIDPLKFPATTAWMTNFLSHPVIKDTLPPRDKMID 209
G++DIA ++++++ +++E+ +H+ KFP W +FL+HPV+K++LPPRD +
Sbjct: 178 FGFVDIAAVFVAFYVPLFQEIAGLHLFTAEKFPQLYDWSQDFLNHPVVKESLPPRDPLFA 357
Query: 210 YFHGRKKDLS 219
+F GR + L+
Sbjct: 358 FFKGRYESLT 387
>AV423845
Length = 259
Score = 99.4 bits (246), Expect = 5e-22
Identities = 49/81 (60%), Positives = 59/81 (72%)
Frame = +3
Query: 3 SQDVKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGH 62
S ++KLLS W SPF+ RV AL LKG++YE +EE + KS LLL+ NPVHKKIPVL+H
Sbjct: 9 SNELKLLSGWFSPFAMRVRIALNLKGLDYEVLEETLNPKSDLLLQSNPVHKKIPVLIHKD 188
Query: 63 KTIAESFIIIEYIDETWKQYP 83
K I ES II+EYIDE W P
Sbjct: 189KPICESAIIVEYIDEVWNDGP 251
>TC9907 weakly similar to UP|Q84T17 (Q84T17) Glutathione S-transferase,
partial (44%)
Length = 559
Score = 90.1 bits (222), Expect = 3e-19
Identities = 39/96 (40%), Positives = 66/96 (68%)
Frame = +2
Query: 119 DEQEKALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDP 178
+E+EK + E+ EA++ +E+ ++ K FFGGE IG +DIA ++++W+ + EE ++
Sbjct: 23 EEREKGIQESVEALQFLEKELKHK-FFGGETIGLVDIAGAFLAFWLPVIEESTGFSLLTS 199
Query: 179 LKFPATTAWMTNFLSHPVIKDTLPPRDKMIDYFHGR 214
KFP W +F++HPV+K+ LPPRDK++ +F GR
Sbjct: 200 EKFPKLYKWSQDFINHPVVKENLPPRDKLLGFFKGR 307
>AV779654
Length = 488
Score = 77.4 bits (189), Expect = 2e-15
Identities = 31/93 (33%), Positives = 59/93 (63%)
Frame = -2
Query: 119 DEQEKALNEAREAMEKIEEVIEGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDP 178
+E+EK + E +A+ +E ++ ++FFGGE G++D+A +I++ I +++E+ + +
Sbjct: 484 EEREKKIEELSDALPFLETELKDQKFFGGEEFGFVDLAAVFIAFHIPLFQEIAGLPLFTA 305
Query: 179 LKFPATTAWMTNFLSHPVIKDTLPPRDKMIDYF 211
KFP W FL+HPV+ +++PPRD + F
Sbjct: 304 EKFPKLYQWSPAFLNHPVVTESMPPRDPLFSAF 206
>AV765173
Length = 565
Score = 73.9 bits (180), Expect = 2e-14
Identities = 38/118 (32%), Positives = 61/118 (51%), Gaps = 5/118 (4%)
Frame = -3
Query: 98 WANFTEQKLLETVFVAMCITGDEQEKAL-NEAREAMEKIEEVI----EGKRFFGGENIGY 152
W + + K ++ + DE ++ L + E + ++E+V EGK FFGGE IG+
Sbjct: 563 WGAYIDDKWFLSMRDMLGAQDDEAKEPLFAQMEEVLGRLEDVFNKCSEGKAFFGGETIGF 384
Query: 153 LDIAVGWISYWIHIWEEVGSIHIIDPLKFPATTAWMTNFLSHPVIKDTLPPRDKMIDY 210
+DIA G W+ + EE+ + D KFPA W F + P +K LP +K+I +
Sbjct: 383 IDIAFGCYLTWLSVTEEMTGRKVFDEAKFPALVKWAATFAADPAVKGVLPETEKLIGF 210
>TC10564 similar to UP|Q9FQD3 (Q9FQD3) Glutathione S-transferase GST 25 ,
partial (53%)
Length = 500
Score = 66.6 bits (161), Expect = 3e-12
Identities = 39/92 (42%), Positives = 56/92 (60%), Gaps = 3/92 (3%)
Frame = +1
Query: 6 VKLLSFWPSPFSKRVEWALKLKGVEYEYVEEDIFN---KSSLLLELNPVHKKIPVLVHGH 62
+KL S+W S S RV AL LKG++YEY ++ + L+LNPV +PVLV G
Sbjct: 151 LKLYSYWRSSCSFRVRIALNLKGLKYEYKPVNLLKGEQSNPEFLKLNPVGC-VPVLVDGP 327
Query: 63 KTIAESFIIIEYIDETWKQYPLLPHDPYERAL 94
I +SF II Y+++ + +PLLP D +RA+
Sbjct: 328 AVIFDSFAIIMYLEDKYPNHPLLPRDINQRAI 423
>TC16057 weakly similar to UP|Q9M6R4 (Q9M6R4) Glutathione S-transferase,
partial (37%)
Length = 590
Score = 63.2 bits (152), Expect = 4e-11
Identities = 30/96 (31%), Positives = 49/96 (50%), Gaps = 4/96 (4%)
Frame = +3
Query: 119 DEQEKALNEAREAMEKIEEVI----EGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIH 174
DE+ K + RE + +E+ +G FFGG+ IGYLDIA G W+ + E +
Sbjct: 60 DERNKRIPAVREGLLLLEDAFPKSSQGHDFFGGDPIGYLDIAFGSFLGWLRVVEVSHRVK 239
Query: 175 IIDPLKFPATTAWMTNFLSHPVIKDTLPPRDKMIDY 210
+DP P W F +H ++D +P +K+ ++
Sbjct: 240 FLDPENTPGLVKWAEKFCAHIAVQDIMPETEKLHEF 347
>TC13189 similar to UP|Q9FUS9 (Q9FUS9) Glutathione S-transferase, partial
(27%)
Length = 544
Score = 56.2 bits (134), Expect = 5e-09
Identities = 25/69 (36%), Positives = 38/69 (54%)
Frame = +2
Query: 140 EGKRFFGGENIGYLDIAVGWISYWIHIWEEVGSIHIIDPLKFPATTAWMTNFLSHPVIKD 199
+GK FFGG+ IGYLDIA G W+ E+ ++ K PA W F++ P +K
Sbjct: 59 KGKAFFGGDKIGYLDIAFGSFLGWLSAIEQEYQRKVLVESKAPALVKWAERFVADPAVKG 238
Query: 200 TLPPRDKMI 208
+P D+++
Sbjct: 239 IIPETDRVV 265
>TC17380 similar to UP|Q84UH4 (Q84UH4) Dehydroascorbate reductase, complete
Length = 875
Score = 44.7 bits (104), Expect = 1e-05
Identities = 34/143 (23%), Positives = 69/143 (47%)
Frame = +2
Query: 15 PFSKRVEWALKLKGVEYEYVEEDIFNKSSLLLELNPVHKKIPVLVHGHKTIAESFIIIEY 74
PFS+RV L+ K + ++ D+ NK L++NP K+PV+ G K +A+S +I+
Sbjct: 149 PFSQRVLLTLEEKKIPHKIHLIDLSNKPQWFLDVNP-EGKVPVVKFGDKWVADSDVIVGI 325
Query: 75 IDETWKQYPLLPHDPYERALARFWANFTEQKLLETVFVAMCITGDEQEKALNEAREAMEK 134
++E + + L+ + ++ + +F F+ D E+AL A++
Sbjct: 326 LEEKYPEPSLVTPPEFASVGSKIFGSFVS-------FLKSKDANDGTEQALVAELSALD- 481
Query: 135 IEEVIEGKRFFGGENIGYLDIAV 157
E + + GE + +D+++
Sbjct: 482 -EHLKAHGPYVAGERVTAVDLSL 547
>TC12677 similar to UP|Q9FQD6 (Q9FQD6) Glutathione S-transferase GST 22
(Fragment) , complete
Length = 876
Score = 41.6 bits (96), Expect = 1e-04
Identities = 42/155 (27%), Positives = 65/155 (41%), Gaps = 16/155 (10%)
Frame = +3
Query: 18 KRVEWALKLKGVEYEYVEEDIF---NKSSLLLELNPVHKKIPVLVHGHKTIAESFIIIEY 74
KRV L K +E+E V DIF +K L++ P ++PV+ G T+ ES II Y
Sbjct: 69 KRVMVCLIEKEIEFETVPVDIFKREHKDPEYLKIQP-FGELPVIQDGDYTLYESRAIIRY 245
Query: 75 IDETWKQY--PLLPHDPYERALARFWANFTE-------QKLLETVFVAMCITGDEQEKAL 125
E +K LL ER L W + L+ V A + K +
Sbjct: 246 YAEKYKNQGTELLGKTIEERGLVEQWLDVEAHSFQPAIHNLVNHVLFAPILGTPSDPKVI 425
Query: 126 NEAREAMEKI----EEVIEGKRFFGGENIGYLDIA 156
E+ E + K+ EE + ++ G+ D++
Sbjct: 426 EESDEKLVKVLDIYEERLSKSKYLAGDFFSLADLS 530
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.140 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,606,831
Number of Sequences: 28460
Number of extensions: 67412
Number of successful extensions: 359
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 350
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 352
length of query: 228
length of database: 4,897,600
effective HSP length: 87
effective length of query: 141
effective length of database: 2,421,580
effective search space: 341442780
effective search space used: 341442780
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0162b.4


