

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0158.2
(125 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
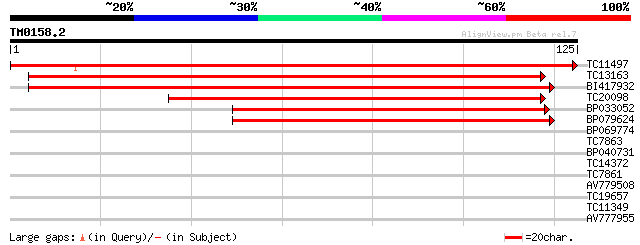
Score E
Sequences producing significant alignments: (bits) Value
TC11497 similar to UP|Q8S925 (Q8S925) Autophagy 8h (Symbiosis-re... 238 2e-64
TC13163 similar to UP|APG8_YEAST (P38182) Autophagy protein 8 [C... 135 2e-33
BI417932 124 4e-30
TC20098 similar to UP|APG8_YEAST (P38182) Autophagy protein 8 [C... 108 2e-25
BP033052 93 1e-20
BP079624 82 2e-17
BP069774 27 0.77
TC7863 similar to UP|AAR86686 (AAR86686) Proline dehydrogenase ... 26 2.3
BP040731 26 2.3
TC14372 similar to UP|Q8L784 (Q8L784) Cytoplasmic aconitate hydr... 25 3.8
TC7861 similar to UP|AAR86686 (AAR86686) Proline dehydrogenase ... 25 3.8
AV779508 25 5.0
TC19657 25 5.0
TC11349 similar to UP|Q9SIN1 (Q9SIN1) Expressed protein (At2g425... 24 6.6
AV777955 24 6.6
>TC11497 similar to UP|Q8S925 (Q8S925) Autophagy 8h (Symbiosis-related like
protein), partial (88%)
Length = 584
Score = 238 bits (608), Expect = 2e-64
Identities = 125/155 (80%), Positives = 125/155 (80%), Gaps = 30/155 (19%)
Frame = +3
Query: 1 MGGSSSFKDEFTF------------------------------DQRLEESREIIAKYPDR 30
MGGSSSFKDEFTF DQRLEESREIIAKYPDR
Sbjct: 18 MGGSSSFKDEFTFGALNSSSPFLPISLT*FTHHCLSLFDFARADQRLEESREIIAKYPDR 197
Query: 31 VPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIHILSSRLSLPPGKALFVFVKNTLPQTA 90
VPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIHILSSRLSLPPGKALFVFVKNTLPQTA
Sbjct: 198 VPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIHILSSRLSLPPGKALFVFVKNTLPQTA 377
Query: 91 SMMDSVYRSFRDEDGFLYMYYSTEKTFGSVHNINC 125
SMMDSVYRSFRDEDGFLYMYYSTEKTFGSVHNINC
Sbjct: 378 SMMDSVYRSFRDEDGFLYMYYSTEKTFGSVHNINC 482
>TC13163 similar to UP|APG8_YEAST (P38182) Autophagy protein 8 [Contains:
Apg8FG], partial (76%)
Length = 490
Score = 135 bits (341), Expect = 2e-33
Identities = 64/114 (56%), Positives = 84/114 (73%)
Frame = +1
Query: 5 SSFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIH 64
SS K E ++R E+ I KYPDR+PVIVER KSD+P++ K+KYLVP DL+VG F++
Sbjct: 76 SSSKMEHPLERRQAEAARIREKYPDRIPVIVERAEKSDVPDIDKKKYLVPADLTVGQFVY 255
Query: 65 ILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFG 118
++ R+ L P KA+F+FVKN LP TA+MM ++Y +DEDGFLYM YS E TFG
Sbjct: 256 VVRKRIKLSPEKAIFIFVKNILPPTAAMMSAIYEENKDEDGFLYMTYSGENTFG 417
>BI417932
Length = 503
Score = 124 bits (312), Expect = 4e-30
Identities = 57/116 (49%), Positives = 81/116 (69%)
Frame = +3
Query: 5 SSFKDEFTFDQRLEESREIIAKYPDRVPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIH 64
S FK E ++R E+ I KYPDR+PVIVE+ +SD+P + K+KYLVP DL+VG F++
Sbjct: 120 SYFKQEHDLEKRRAEAVRIREKYPDRIPVIVEKAERSDIPSIDKKKYLVPADLTVGQFVY 299
Query: 65 ILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYMYYSTEKTFGSV 120
++ R+ L KA+F+FV N LP T ++M ++Y +DEDGFLY+ YS E TFG +
Sbjct: 300 VIRKRIKLSAEKAIFIFVDNVLPPTGAIMSAIYDEKKDEDGFLYVTYSGENTFGDL 467
>TC20098 similar to UP|APG8_YEAST (P38182) Autophagy protein 8 [Contains:
Apg8FG], partial (71%)
Length = 468
Score = 108 bits (271), Expect = 2e-25
Identities = 49/83 (59%), Positives = 64/83 (77%)
Frame = +1
Query: 36 ERYLKSDLPELGKRKYLVPRDLSVGHFIHILSSRLSLPPGKALFVFVKNTLPQTASMMDS 95
ER KSD+PE+ K+KYLVP DL+VG F++++ R+ L P KA+F+FVKN LP TA+MM +
Sbjct: 1 ERAEKSDVPEIDKKKYLVPADLTVGQFVYVVRKRIKLSPEKAIFIFVKNILPPTAAMMSA 180
Query: 96 VYRSFRDEDGFLYMYYSTEKTFG 118
+Y +DEDGFLYM YS E TFG
Sbjct: 181 IYEENKDEDGFLYMTYSGENTFG 249
>BP033052
Length = 469
Score = 93.2 bits (230), Expect = 1e-20
Identities = 42/70 (60%), Positives = 54/70 (77%)
Frame = -1
Query: 50 KYLVPRDLSVGHFIHILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYM 109
+YLVP DL+VG F++++ R+ L KA+FVF+KNTLP TAS+M +VY +DEDGFLYM
Sbjct: 307 RYLVPADLTVGQFVYVVRKRIKLSAEKAIFVFIKNTLPPTASLMSAVYEENKDEDGFLYM 128
Query: 110 YYSTEKTFGS 119
YS E TFGS
Sbjct: 127 TYSGENTFGS 98
>BP079624
Length = 456
Score = 82.4 bits (202), Expect = 2e-17
Identities = 35/71 (49%), Positives = 51/71 (71%)
Frame = -3
Query: 50 KYLVPRDLSVGHFIHILSSRLSLPPGKALFVFVKNTLPQTASMMDSVYRSFRDEDGFLYM 109
+YLVP DL+VG F++++ R+ L KA+F+FV N LP T ++M ++Y +DEDGFLY+
Sbjct: 370 RYLVPADLTVGQFVYVIRKRIKLSAEKAIFIFVDNVLPPTGAIMSAIYDEKKDEDGFLYV 191
Query: 110 YYSTEKTFGSV 120
YS E TFG +
Sbjct: 190 TYSGENTFGDL 158
>BP069774
Length = 446
Score = 27.3 bits (59), Expect = 0.77
Identities = 19/65 (29%), Positives = 30/65 (45%)
Frame = -3
Query: 31 VPVIVERYLKSDLPELGKRKYLVPRDLSVGHFIHILSSRLSLPPGKALFVFVKNTLPQTA 90
+P++++ + + LP L VPR L HFI S S+ GK+ P
Sbjct: 201 IPLVIKSQI-TQLPSLHPAARRVPRRLKAMHFIQPYSKYSSITSGKS---------PCNG 52
Query: 91 SMMDS 95
SMM++
Sbjct: 51 SMMEA 37
>TC7863 similar to UP|AAR86686 (AAR86686) Proline dehydrogenase ,
partial (68%)
Length = 1326
Score = 25.8 bits (55), Expect = 2.3
Identities = 12/30 (40%), Positives = 15/30 (50%)
Frame = +1
Query: 56 DLSVGHFIHILSSRLSLPPGKALFVFVKNT 85
D + F+H + SLPP FV VK T
Sbjct: 25 DRNANGFLHTVDVSRSLPPSSVSFVIVKIT 114
>BP040731
Length = 492
Score = 25.8 bits (55), Expect = 2.3
Identities = 18/42 (42%), Positives = 21/42 (49%)
Frame = -1
Query: 36 ERYLKSDLPELGKRKYLVPRDLSVGHFIHILSSRLSLPPGKA 77
ER S LP LG+ PRD + GH H L R P G+A
Sbjct: 459 ERTTTSTLP-LGRSSSPSPRDATAGHHHHRL-RRAKAPRGEA 340
>TC14372 similar to UP|Q8L784 (Q8L784) Cytoplasmic aconitate hydratase
(At2g05710), partial (7%)
Length = 546
Score = 25.0 bits (53), Expect = 3.8
Identities = 13/31 (41%), Positives = 16/31 (50%)
Frame = -3
Query: 47 GKRKYLVPRDLSVGHFIHILSSRLSLPPGKA 77
G + YL + H H + SR LPPGKA
Sbjct: 364 GVKNYLRGIEGIFSHIYHKIYSRHILPPGKA 272
>TC7861 similar to UP|AAR86686 (AAR86686) Proline dehydrogenase , partial
(40%)
Length = 943
Score = 25.0 bits (53), Expect = 3.8
Identities = 12/33 (36%), Positives = 17/33 (51%)
Frame = +1
Query: 62 FIHILSSRLSLPPGKALFVFVKNTLPQTASMMD 94
F+H + SLPP FV VK T S+++
Sbjct: 685 FLHTVDVSRSLPPSSVSFVIVKITAICPMSLLE 783
>AV779508
Length = 431
Score = 24.6 bits (52), Expect = 5.0
Identities = 13/31 (41%), Positives = 18/31 (57%), Gaps = 3/31 (9%)
Frame = -3
Query: 25 AKYPDRV---PVIVERYLKSDLPELGKRKYL 52
A Y R+ PV+V+ Y+K+ LP G R L
Sbjct: 288 AGYTSRIRQYPVLVQAYIKAKLPAYGIRDRL 196
>TC19657
Length = 632
Score = 24.6 bits (52), Expect = 5.0
Identities = 14/25 (56%), Positives = 15/25 (60%), Gaps = 2/25 (8%)
Frame = +2
Query: 57 LSVGHFIHIL--SSRLSLPPGKALF 79
LS HFIHIL SS SLP +F
Sbjct: 20 LSPSHFIHILTHSSSFSLPRRHGIF 94
>TC11349 similar to UP|Q9SIN1 (Q9SIN1) Expressed protein
(At2g42580/F14N22.15), partial (5%)
Length = 474
Score = 24.3 bits (51), Expect = 6.6
Identities = 8/21 (38%), Positives = 14/21 (66%)
Frame = +3
Query: 24 IAKYPDRVPVIVERYLKSDLP 44
+ +PDR PV+ R+L+ +P
Sbjct: 387 VLAHPDRSPVVETRFLEDPIP 449
>AV777955
Length = 429
Score = 24.3 bits (51), Expect = 6.6
Identities = 11/29 (37%), Positives = 15/29 (50%)
Frame = +1
Query: 45 ELGKRKYLVPRDLSVGHFIHILSSRLSLP 73
E GKRK PR + HF ++ S+ P
Sbjct: 91 ESGKRKPSKPRSWTWDHFTKVIGSKSKTP 177
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.140 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,855,286
Number of Sequences: 28460
Number of extensions: 20777
Number of successful extensions: 107
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 106
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 107
length of query: 125
length of database: 4,897,600
effective HSP length: 80
effective length of query: 45
effective length of database: 2,620,800
effective search space: 117936000
effective search space used: 117936000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 50 (23.9 bits)
Lotus: description of TM0158.2


