

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0158.11
(361 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
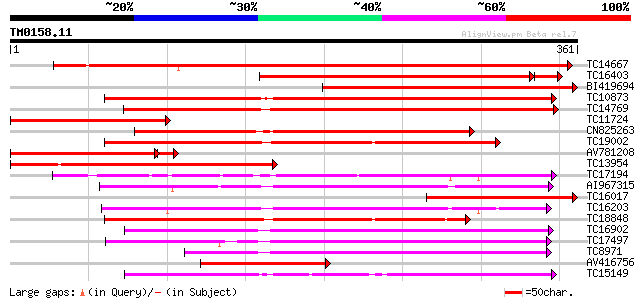
Score E
Sequences producing significant alignments: (bits) Value
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 494 e-141
TC16403 homologue to UP|Q9LKY4 (Q9LKY4) Pti1 kinase-like protein... 318 2e-91
BI419694 310 2e-85
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 248 1e-66
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 224 2e-59
TC11724 weakly similar to UP|Q9M1Q2 (Q9M1Q2) Serine/threonine pr... 215 8e-57
CN825263 215 1e-56
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 212 9e-56
AV781208 203 5e-55
TC13954 similar to UP|Q9LKY3 (Q9LKY3) Pti1 kinase-like protein, ... 206 4e-54
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 202 5e-53
AI967315 201 1e-52
TC16017 homologue to UP|Q9M1Q2 (Q9M1Q2) Serine/threonine protein... 195 1e-50
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 194 3e-50
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 194 3e-50
TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-... 190 3e-49
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 168 1e-42
TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P... 163 4e-41
AV416756 161 2e-40
TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like pro... 147 4e-36
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 494 bits (1272), Expect = e-141
Identities = 254/333 (76%), Positives = 277/333 (82%), Gaps = 3/333 (0%)
Frame = +2
Query: 29 GNDGNHHASETAKQGGQTVKPQPIEVPNISEDELKEVTDNFGQDSLIGEGSYGRVYYGVL 88
GN AS K Q P PIEVP +S DELKE TDNFG +LIGEGSYGRVYY L
Sbjct: 242 GNSKGSKASAPVKHETQKAPP-PIEVPALSLDELKEKTDNFGSKALIGEGSYGRVYYATL 418
Query: 89 KNGQAAAIKKLDASKQPD--EEFLAQVSMVSRLKHENFVQLLGYSVDGNSRILVYEFASN 146
+G A A+KKLD S +P+ EFL QVSMVSRLK++NFV+L GY V+GN R+L YEFA+
Sbjct: 419 NDGNAVAVKKLDVSSEPETNNEFLTQVSMVSRLKNDNFVELHGYCVEGNLRVLAYEFATM 598
Query: 147 GSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVGAARGLEYLHEKADPHIIHRDIKSSNVL 206
GSLHDILHGRKGV+GAQPGP L W QRV+IAV AARGLEYLHEK P IIHRDI+SSNVL
Sbjct: 599 GSLHDILHGRKGVQGAQPGPTLDWIQRVRIAVDAARGLEYLHEKVQPAIIHRDIRSSNVL 778
Query: 207 IFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVL 266
IF+D AKIADF+LSNQAPDMAARLHSTRVLGTFGYHAPEYAMTGQL KSDVYSFGVVL
Sbjct: 779 IFEDYKAKIADFNLSNQAPDMAARLHSTRVLGTFGYHAPEYAMTGQLTQKSDVYSFGVVL 958
Query: 267 LELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKVRQCVDTRLGGEYPPKAVAKMAAVAAL 326
LELLTGRKPVDHT+PRGQQSLVTWATP+LSEDKV+QCVD +L GEYPPK VAK+AAVAAL
Sbjct: 959 LELLTGRKPVDHTMPRGQQSLVTWATPRLSEDKVKQCVDPKLKGEYPPKGVAKLAAVAAL 1138
Query: 327 CVQYEADFRPNMSIVVKALQPLL-TARPGPAGE 358
CVQYEA+FRPNMSIVVKALQPLL T PA E
Sbjct: 1139CVQYEAEFRPNMSIVVKALQPLLKTPAAAPAPE 1237
>TC16403 homologue to UP|Q9LKY4 (Q9LKY4) Pti1 kinase-like protein, partial
(55%)
Length = 876
Score = 318 bits (816), Expect(2) = 2e-91
Identities = 157/175 (89%), Positives = 166/175 (94%)
Frame = +3
Query: 160 KGAQPGPVLTWAQRVKIAVGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFD 219
+GAQPG VL+W QRVKIAVGAARGLEYLHEKA+ HIIHR IKSSN+L+FDDDVAKIADFD
Sbjct: 12 EGAQPGLVLSWTQRVKIAVGAARGLEYLHEKAETHIIHRYIKSSNILLFDDDVAKIADFD 191
Query: 220 LSNQAPDMAARLHSTRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHT 279
LSNQAPD AARLHSTRVLGTFGYHAPEYAMTGQL +KSDVYSFGVVLLELLTGRKPVDHT
Sbjct: 192 LSNQAPDAAARLHSTRVLGTFGYHAPEYAMTGQLTSKSDVYSFGVVLLELLTGRKPVDHT 371
Query: 280 LPRGQQSLVTWATPKLSEDKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADF 334
LPRGQQSLVTWATPKLSEDKV+QCVD RL GEYP K+VAKMAAVAALCVQYEA+F
Sbjct: 372 LPRGQQSLVTWATPKLSEDKVKQCVDARLKGEYPVKSVAKMAAVAALCVQYEAEF 536
Score = 33.9 bits (76), Expect(2) = 2e-91
Identities = 15/18 (83%), Positives = 16/18 (88%)
Frame = +1
Query: 335 RPNMSIVVKALQPLLTAR 352
RPNMSI+VKALQPLL R
Sbjct: 538 RPNMSIIVKALQPLLNTR 591
>BI419694
Length = 623
Score = 310 bits (795), Expect = 2e-85
Identities = 154/162 (95%), Positives = 157/162 (96%)
Frame = +3
Query: 200 IKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTFGYHAPEYAMTGQLNAKSDV 259
+KSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTFGYHAPEYAMTGQLNAKSDV
Sbjct: 24 VKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTFGYHAPEYAMTGQLNAKSDV 203
Query: 260 YSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKVRQCVDTRLGGEYPPKAVAK 319
YSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKVRQCVD RLGGEYPPKAVAK
Sbjct: 204 YSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKVRQCVDARLGGEYPPKAVAK 383
Query: 320 MAAVAALCVQYEADFRPNMSIVVKALQPLLTARPGPAGETAN 361
+AAVAALCVQYEADFRPNMSIVVKALQPLL AR GPAG+ N
Sbjct: 384 LAAVAALCVQYEADFRPNMSIVVKALQPLLNARHGPAGDAPN 509
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 248 bits (633), Expect = 1e-66
Identities = 136/290 (46%), Positives = 194/290 (66%), Gaps = 2/290 (0%)
Frame = +1
Query: 61 ELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLDAS-KQPDEEFLAQVSMVSRL 119
EL + T NF + +LIGEG +G+VY G L G+A A+K+L +Q +EF+ +V M+S L
Sbjct: 427 ELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQEFVMEVLMLSLL 606
Query: 120 KHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVG 179
H N V+L+GY DG+ R+LVYE+ GSL D L K +P L W+ R+K+AVG
Sbjct: 607 HHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDK--EP---LNWSTRMKVAVG 771
Query: 180 AARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGT 239
AARGLEYLH ADP +I+RD+KS+N+L+ ++ K++DF L+ P STRV+GT
Sbjct: 772 AARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGT 951
Query: 240 FGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSE-D 298
+GY APEYAM+G+L KSD+YSFGVVLLELLTGR+ +D + G+Q+LV+WA P S+
Sbjct: 952 YGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRR 1131
Query: 299 KVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPL 348
+ VD L G +P + + + A+ A+C+Q + FRP ++ +V AL+ L
Sbjct: 1132RFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYL 1281
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 224 bits (570), Expect = 2e-59
Identities = 124/278 (44%), Positives = 172/278 (61%), Gaps = 1/278 (0%)
Frame = +3
Query: 73 SLIGEGSYGRVYYGVLKNGQAAAIK-KLDASKQPDEEFLAQVSMVSRLKHENFVQLLGYS 131
+LIGEG +G VY G L +GQ A+K + S Q EF +++++S ++HEN V LLGY
Sbjct: 2175 TLIGEGGFGSVYRGTLNDGQEVAVKVRSSTSTQGTREFDNELNLLSAIQHENLVPLLGYC 2354
Query: 132 VDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVGAARGLEYLHEKA 191
+ + +ILVY F SNGSL D L+G + +L W R+ IA+GAARGL YLH
Sbjct: 2355 NESDQQILVYPFMSNGSLQDRLYGEPAKR-----KILDWPTRLSIALGAARGLAYLHTFP 2519
Query: 192 DPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTFGYHAPEYAMTG 251
+IHRDIKSSN+L+ AK+ADF S AP S V GT GY PEY T
Sbjct: 2520 GRSVIHRDIKSSNILLDHSMCAKVADFGFSKYAPQEGDSYVSLEVRGTAGYLDPEYYKTQ 2699
Query: 252 QLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKVRQCVDTRLGGE 311
QL+ KSDV+SFGVVLLE+++GR+P++ PR + SLV WATP + KV + VD + G
Sbjct: 2700 QLSEKSDVFSFGVVLLEIVSGREPLNIKRPRTEWSLVEWATPYIRGSKVDEIVDPGIKGG 2879
Query: 312 YPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPLL 349
Y +A+ ++ VA C++ + +RP+M +V+ L+ L
Sbjct: 2880 YHAEAMWRVVEVALQCLEPFSTYRPSMVAIVRELEDAL 2993
>TC11724 weakly similar to UP|Q9M1Q2 (Q9M1Q2) Serine/threonine protein
kinase-like protein, partial (20%)
Length = 535
Score = 215 bits (548), Expect = 8e-57
Identities = 102/102 (100%), Positives = 102/102 (100%)
Frame = +1
Query: 1 MSCFNCCEEDDFHKTAESGGPYVVKNPSGNDGNHHASETAKQGGQTVKPQPIEVPNISED 60
MSCFNCCEEDDFHKTAESGGPYVVKNPSGNDGNHHASETAKQGGQTVKPQPIEVPNISED
Sbjct: 229 MSCFNCCEEDDFHKTAESGGPYVVKNPSGNDGNHHASETAKQGGQTVKPQPIEVPNISED 408
Query: 61 ELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLDAS 102
ELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLDAS
Sbjct: 409 ELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLDAS 534
>CN825263
Length = 663
Score = 215 bits (547), Expect = 1e-56
Identities = 116/218 (53%), Positives = 148/218 (67%), Gaps = 1/218 (0%)
Frame = +1
Query: 80 YGRVYYGVLKNGQAAAIKKLDASKQPD-EEFLAQVSMVSRLKHENFVQLLGYSVDGNSRI 138
+G VY G+L +G+ A+K L Q EFLA+V M+SRL H N V+L+G ++ +R
Sbjct: 4 FGLVYKGILNDGRDVAVKILKRDDQRGGREFLAEVEMLSRLHHRNLVKLIGICIEKQTRC 183
Query: 139 LVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVGAARGLEYLHEKADPHIIHR 198
L+YE NGS+ LHG + GP L W R+KIA+GAARGL YLHE ++P +IHR
Sbjct: 184 LIYELVPNGSVESHLHGAD----KETGP-LDWNARMKIALGAARGLAYLHEDSNPCVIHR 348
Query: 199 DIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTFGYHAPEYAMTGQLNAKSD 258
D KSSN+L+ D K++DF L+ A D + ST V+GTFGY APEYAMTG L KSD
Sbjct: 349 DFKSSNILLECDFTPKVSDFGLARTALDEGNKHISTHVMGTFGYLAPEYAMTGHLLVKSD 528
Query: 259 VYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLS 296
VYS+GVVLLELLTG KPVD + P GQ++LVTWA P L+
Sbjct: 529 VYSYGVVLLELLTGTKPVDLSQPPGQENLVTWARPILT 642
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 212 bits (539), Expect = 9e-56
Identities = 110/253 (43%), Positives = 160/253 (62%), Gaps = 1/253 (0%)
Frame = +1
Query: 61 ELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLDA-SKQPDEEFLAQVSMVSRL 119
EL T+NF D+ +GEG +G VY+G L +G A+K+L S + D EF +V +++R+
Sbjct: 40 ELHSATNNFNYDNKLGEGGFGSVYWGQLWDGSQIAVKRLKVWSNKADMEFAVEVEILARV 219
Query: 120 KHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVG 179
+H+N + L GY +G R++VY++ N SL LHG+ + +L W +R+ IA+G
Sbjct: 220 RHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHGQHSSEC-----LLDWNRRMNIAIG 384
Query: 180 AARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGT 239
+A G+ YLH +A PHIIHRDIK+SNVL+ D A++ADF + PD A + +TRV GT
Sbjct: 385 SAEGIVYLHHQATPHIIHRDIKASNVLLDSDFQARVADFGFAKLIPDGATHV-TTRVKGT 561
Query: 240 FGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDK 299
GY APEYAM G+ N DV+SFG++LLEL +G+KP++ ++S+ WA P K
Sbjct: 562 LGYLAPEYAMLGKANECCDVFSFGILLLELASGKKPLEKLSSTVKRSINDWALPLACAKK 741
Query: 300 VRQCVDTRLGGEY 312
+ D RL GEY
Sbjct: 742 FTEFADPRLNGEY 780
>AV781208
Length = 524
Score = 203 bits (516), Expect(2) = 5e-55
Identities = 95/95 (100%), Positives = 95/95 (100%)
Frame = -2
Query: 1 MSCFNCCEEDDFHKTAESGGPYVVKNPSGNDGNHHASETAKQGGQTVKPQPIEVPNISED 60
MSCFNCCEEDDFHKTAESGGPYVVKNPSGNDGNHHASETAKQGGQTVKPQPIEVPNISED
Sbjct: 322 MSCFNCCEEDDFHKTAESGGPYVVKNPSGNDGNHHASETAKQGGQTVKPQPIEVPNISED 143
Query: 61 ELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAA 95
ELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAA
Sbjct: 142 ELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAA 38
Score = 28.1 bits (61), Expect(2) = 5e-55
Identities = 12/13 (92%), Positives = 13/13 (99%)
Frame = -1
Query: 95 AIKKLDASKQPDE 107
+IKKLDASKQPDE
Sbjct: 41 SIKKLDASKQPDE 3
>TC13954 similar to UP|Q9LKY3 (Q9LKY3) Pti1 kinase-like protein, partial
(47%)
Length = 553
Score = 206 bits (525), Expect = 4e-54
Identities = 101/170 (59%), Positives = 126/170 (73%)
Frame = +2
Query: 1 MSCFNCCEEDDFHKTAESGGPYVVKNPSGNDGNHHASETAKQGGQTVKPQPIEVPNISED 60
M CF C+ +D + TA+ GGP++ P+GN+ ++H T T+ QPI VP++S D
Sbjct: 47 MGCFGFCKGNDAYTTADKGGPFMQTYPTGNN-SYHGRHTPIITPPTINFQPIAVPSLSID 223
Query: 61 ELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLDASKQPDEEFLAQVSMVSRLK 120
ELK VTDNFG S IGEG+YG+VY LKNG+ AIKKL +SKQPD+E L+QVS+VSRLK
Sbjct: 224 ELKSVTDNFGSKSFIGEGAYGKVYRATLKNGREVAIKKLHSSKQPDQELLSQVSVVSRLK 403
Query: 121 HENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTW 170
HEN V+L+ Y VDG R L YE+A NGSLHDI+HGRKGVKGAQPG VL+W
Sbjct: 404 HENVVELVSYCVDGPCRALAYEYAPNGSLHDIIHGRKGVKGAQPGLVLSW 553
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 202 bits (515), Expect = 5e-53
Identities = 132/327 (40%), Positives = 184/327 (55%), Gaps = 6/327 (1%)
Frame = +1
Query: 28 SGNDGNHHASETAKQGGQTVKPQPIEVPNISEDELKEVTDNFGQDSLIGEGSYGRVYYGV 87
SG+ G AS T K S EL + T+NF D+ IG+G +G VYY
Sbjct: 979 SGSSGPGTASATGLTSIMVAKSM-----EFSYQELAKATNNFSLDNKIGQGGFGAVYYAE 1143
Query: 88 LKNGQAAAIKKLDASKQPDEEFLAQVSMVSRLKHENFVQLLGYSVDGNSRILVYEFASNG 147
L+ G+ AIKK+D Q EFL ++ +++ + H N V+L+GY V+G S LVYE NG
Sbjct: 1144 LR-GKKTAIKKMDV--QASTEFLCELKVLTHVHHLNLVRLIGYCVEG-SLFLVYEHIDNG 1311
Query: 148 SLHDILHGRKGVKGAQPGPVLTWAQRVKIAVGAARGLEYLHEKADPHIIHRDIKSSNVLI 207
+L LHG G +P P W+ RV+IA+ AARGLEY+HE P IHRD+KS+N+LI
Sbjct: 1312 NLGQYLHG----SGKEPLP---WSSRVQIALDAARGLEYIHEHTVPVYIHRDVKSANILI 1470
Query: 208 FDDDVAKIADFDLSNQAPDMAARLHSTRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLL 267
+ K+ADF L+ + ++ TR++GTFGY PEYA G ++ K DVY+FGVVL
Sbjct: 1471 DKNLRGKVADFGLT-KLIEVGNSTLQTRLVGTFGYMPPEYAQYGDISPKIDVYAFGVVLF 1647
Query: 268 ELLTGRKPVDHT--LPRGQQSLVTWATPKLSE----DKVRQCVDTRLGGEYPPKAVAKMA 321
EL++ + V T L + LV L++ D +R+ VD RLG YP +V K+A
Sbjct: 1648 ELISAKNAVLKTGELVAESKGLVALFEEALNKSDPCDALRKLVDPRLGENYPIDSVLKIA 1827
Query: 322 AVAALCVQYEADFRPNMSIVVKALQPL 348
+ C + RP+M +V AL L
Sbjct: 1828 QLGRACTRDNPLLRPSMRSLVVALMTL 1908
>AI967315
Length = 1308
Score = 201 bits (512), Expect = 1e-52
Identities = 111/292 (38%), Positives = 174/292 (59%), Gaps = 3/292 (1%)
Frame = +1
Query: 58 SEDELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLDAS---KQPDEEFLAQVS 114
S +EL T+ F ++++G+G Y VY G L++G A+K+L + ++ ++EFL ++
Sbjct: 115 SYEELFHATNGFSSENMVGKGGYAEVYKGRLESGDEIAVKRLTRTCRDERKEKEFLTEIG 294
Query: 115 MVSRLKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRV 174
+ + H N + LLG +D N LV+E ++ GS+ ++H K L W R
Sbjct: 295 TIGHVCHSNVMPLLGCCID-NGLYLVFELSTVGSVASLIHDEKMAP-------LDWKTRY 450
Query: 175 KIAVGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHST 234
KI +G ARGL YLH+ IIHRDIK+SN+L+ +D +I+DF L+ P
Sbjct: 451 KIVLGTARGLHYLHKGCQRRIIHRDIKASNILLTEDFEPQISDFGLAKWLPSQWTHHSIA 630
Query: 235 RVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPK 294
+ GTFG+ APEY M G ++ K+DV++FGV LLE+++GRKPVD + QSL TWA P
Sbjct: 631 PIEGTFGHLAPEYYMHGVVDEKTDVFAFGVFLLEVISGRKPVDGS----HQSLHTWAKPI 798
Query: 295 LSEDKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQ 346
LS+ ++ + VD RL G Y ++A A+LC++ + +RP MS V++ ++
Sbjct: 799 LSKWEIEKLVDPRLEGCYDVTQFNRVAFAASLCIRASSTWRPTMSEVLEVME 954
>TC16017 homologue to UP|Q9M1Q2 (Q9M1Q2) Serine/threonine protein
kinase-like protein, partial (26%)
Length = 629
Score = 195 bits (495), Expect = 1e-50
Identities = 96/96 (100%), Positives = 96/96 (100%)
Frame = +2
Query: 266 LLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKVRQCVDTRLGGEYPPKAVAKMAAVAA 325
LLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKVRQCVDTRLGGEYPPKAVAKMAAVAA
Sbjct: 2 LLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKVRQCVDTRLGGEYPPKAVAKMAAVAA 181
Query: 326 LCVQYEADFRPNMSIVVKALQPLLTARPGPAGETAN 361
LCVQYEADFRPNMSIVVKALQPLLTARPGPAGETAN
Sbjct: 182 LCVQYEADFRPNMSIVVKALQPLLTARPGPAGETAN 289
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 194 bits (492), Expect = 3e-50
Identities = 120/296 (40%), Positives = 168/296 (56%), Gaps = 9/296 (3%)
Frame = +2
Query: 59 EDELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKL--DASKQPDEEFLAQVSMV 116
E + ++V + ++++IG+G G VY G + NG AIK+L S + D F A++ +
Sbjct: 2162 EIKAEDVVECLKEENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYGFRAEIETL 2341
Query: 117 SRLKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKI 176
+++H N ++LLGY + ++ +L+YE+ NGSL + LHG KG L W R KI
Sbjct: 2342 GKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKGGH-------LRWEMRYKI 2500
Query: 177 AVGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRV 236
AV AARGL Y+H P IIHRD+KS+N+L+ D A +ADF L+ D A + +
Sbjct: 2501 AVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSI 2680
Query: 237 LGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLS 296
G++GY APEYA T +++ KSDVYSFGVVLLEL+ GRKPV +V W +S
Sbjct: 2681 AGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEF--GDGVDIVGWVNKTMS 2854
Query: 297 E-------DKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKAL 345
E V VD RL G YP +V M +A +CV+ RP M VV L
Sbjct: 2855 ELSQPSDTALVLAVVDPRLSG-YPLTSVIHMFNIAMMCVKEMGPARPTMREVVHML 3019
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 194 bits (492), Expect = 3e-50
Identities = 105/235 (44%), Positives = 149/235 (62%), Gaps = 2/235 (0%)
Frame = +2
Query: 61 ELKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKLDASKQPDE-EFLAQVSMVSRL 119
EL T F D+ +GEG +G VY+G +G A+KKL A E EF +V ++ R+
Sbjct: 278 ELHAATGGFSDDNKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEVEVLGRV 457
Query: 120 KHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVG 179
+H+N + L GY V + R++VY++ N SL LHG+ V+ L W +R+KIA+G
Sbjct: 458 RHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQFAVEVQ-----LNWQKRMKIAIG 622
Query: 180 AARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGT 239
+A G+ YLH + PHIIHRDIK+SNVL+ D +ADF + P+ + + +TRV GT
Sbjct: 623 SAEGILYLHHEVTPHIIHRDIKASNVLLNSDFEPLVADFGFAKLIPEGVSHM-TTRVKGT 799
Query: 240 FGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVT-WATP 293
GY APEYAM G+++ DVYSFG++LLEL+TGRKP++ LP G + +T WA P
Sbjct: 800 LGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIE-KLPGGVKRTITEWAEP 961
>TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like
protein, partial (46%)
Length = 941
Score = 190 bits (483), Expect = 3e-49
Identities = 110/274 (40%), Positives = 160/274 (58%), Gaps = 1/274 (0%)
Frame = +3
Query: 74 LIGEGSYGRVYYGVLKNGQAAAIKKLDA-SKQPDEEFLAQVSMVSRLKHENFVQLLGYSV 132
L+G G +G+VYYG + G AIK+ + S+Q EF ++ M+S+L+H + V L+GY
Sbjct: 12 LLGVGGFGKVYYGEVDGGTKVAIKRGNPLSEQGVHEFQTEIEMLSKLRHRHLVSLIGYCE 191
Query: 133 DGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVGAARGLEYLHEKAD 192
+ ILVY+ + G+L + L+ + P L W QR++I +GAARGL YLH A
Sbjct: 192 ENTEMILVYDHMAYGTLREHLYKTQK-------PPLPWKQRLEICIGAARGLHYLHTGAK 350
Query: 193 PHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTFGYHAPEYAMTGQ 252
IIHRD+K++N+L+ + VAK++DF LS P + ST V G+FGY PEY Q
Sbjct: 351 YTIIHRDVKTTNILLDEKWVAKVSDFGLSKTGPTLDNTHVSTVVKGSFGYLDPEYFRRQQ 530
Query: 253 LNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKVRQCVDTRLGGEY 312
L KSDVYSFGVVL E+L R ++ +L + Q SL WA+ ++ + Q +D L G+
Sbjct: 531 LTDKSDVYSFGVVLFEILCARPALNPSLAKEQVSLAEWASHCYNKGILDQILDPYLKGKI 710
Query: 313 PPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQ 346
P+ K A A CV + RP+M V+ L+
Sbjct: 711 APECFKKFAETAMKCVSDQGIERPSMGDVLWNLE 812
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 168 bits (426), Expect = 1e-42
Identities = 103/287 (35%), Positives = 161/287 (55%), Gaps = 3/287 (1%)
Frame = +1
Query: 62 LKEVTDNFGQDSLIGEGSYGRVYYGVLKNGQAAAIKKL-DASKQPDEEFLAQVSMVSRLK 120
+ T++F + +GEG +G VY G+L NGQ A+K+L + S Q EEF ++ +++RL+
Sbjct: 1651 ISSATNHFSLSNKLGEGGFGPVYKGLLANGQEIAVKRLSNTSGQGMEEFKNEIKLIARLQ 1830
Query: 121 HENFVQLLGYSV--DGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAV 178
H N V+L G SV D NS +N + +L + ++ W +R++I
Sbjct: 1831 HRNLVKLFGCSVHQDENSH-------ANKKMKILLDSTRS-------KLVDWNKRLQIID 1968
Query: 179 GAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLG 238
G ARGL YLH+ + IIHRD+K+SN+L+ D+ KI+DF L+ + RV+G
Sbjct: 1969 GIARGLLYLHQDSRLRIIHRDLKTSNILLDDEMNPKISDFGLARIFIGDQVEARTKRVMG 2148
Query: 239 TFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSED 298
T+GY PEYA+ G + KSDV+SFGV++LE+++G+K P +L++ A E+
Sbjct: 2149 TYGYMPPEYAVHGSFSIKSDVFSFGVIVLEIISGKKIGRFYDPHHHLNLLSHAWRLWIEE 2328
Query: 299 KVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKAL 345
+ + VD L P + + VA LCVQ + RP+M +V L
Sbjct: 2329 RPLELVDELLDDPVIPTEILRYIHVALLCVQRRPENRPDMLSIVLML 2469
>TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P14.15
{Arabidopsis thaliana;}, partial (37%)
Length = 1079
Score = 163 bits (413), Expect = 4e-41
Identities = 93/235 (39%), Positives = 136/235 (57%), Gaps = 1/235 (0%)
Frame = +3
Query: 112 QVSMVSRLKHENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWA 171
+V +++++ H N V+LLG+ GN RIL+ E+ NG+L + L G +G +L +
Sbjct: 6 EVELLAKIDHRNLVKLLGFIDKGNERILITEYVPNGTLREHLDGLRG-------KILDFN 164
Query: 172 QRVKIAVGAARGLEYLHEKADPHIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARL 231
QR++IA+ A GL YLH A+ IIHRD+KSSN+L+ + AK+ADF + P +
Sbjct: 165 QRLEIAIDVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFARLGPVNGDQT 344
Query: 232 H-STRVLGTFGYHAPEYAMTGQLNAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTW 290
H ST+V GT GY PEY T QL KSDVYSFG++LLE+LTGR+PV+ ++ + W
Sbjct: 345 HISTKVKGTVGYLDPEYMKTHQLTPKSDVYSFGILLLEILTGRRPVELKKAADERVTLRW 524
Query: 291 ATPKLSEDKVRQCVDTRLGGEYPPKAVAKMAAVAALCVQYEADFRPNMSIVVKAL 345
A K +E V + +D + + KM ++ C RPNM V + L
Sbjct: 525 AFRKYNEGSVVELMDPLMEEAVNADVLMKMLDLSFQCAAPIRTDRPNMKSVGEQL 689
>AV416756
Length = 251
Score = 161 bits (407), Expect = 2e-40
Identities = 77/83 (92%), Positives = 80/83 (95%)
Frame = +3
Query: 122 ENFVQLLGYSVDGNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVGAA 181
+NFVQLLGY VDGNSRIL YEFASNGSLHDILHGRKGVKG+QPGPVLTWAQRVKIAV AA
Sbjct: 3 DNFVQLLGYCVDGNSRILAYEFASNGSLHDILHGRKGVKGSQPGPVLTWAQRVKIAVRAA 182
Query: 182 RGLEYLHEKADPHIIHRDIKSSN 204
+GLEYLHEKADPHIIHRDIKSSN
Sbjct: 183 KGLEYLHEKADPHIIHRDIKSSN 251
>TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like protein kinase
protein (AT3g17840/MEB5_6), partial (45%)
Length = 1489
Score = 147 bits (370), Expect = 4e-36
Identities = 92/276 (33%), Positives = 149/276 (53%), Gaps = 1/276 (0%)
Frame = +3
Query: 74 LIGEGSYGRVYYGVLKNGQAAAIKKLDASKQPDEEFLAQVSMVSRLKHENFVQLLGYSVD 133
++G+G++G Y VL+ G A+K+L ++EF ++ V + H++ V L Y
Sbjct: 291 VLGKGTFGTAYKAVLETGLVVAVKRLKDVTISEKEFKDKIETVGAMDHQSLVPLRAYYFS 470
Query: 134 GNSRILVYEFASNGSLHDILHGRKGVKGAQPGPVLTWAQRVKIAVGAARGLEYLHEKADP 193
+ ++LVY++ GSL +LHG KG G P L W R IA+GAARG+EYLH + P
Sbjct: 471 RDEKLLVYDYMPMGSLSALLHGNKGA-GRTP---LNWEIRSGIALGAARGIEYLHSQG-P 635
Query: 194 HIIHRDIKSSNVLIFDDDVAKIADFDLSNQAPDMAARLHSTRVLGTFGYHAPEYAMTGQL 253
++ H +IK+SN+L+ AK++DF L++ + RV GY APE ++
Sbjct: 636 NVSHGNIKASNILLTKSYEAKVSDFGLAHLVGPSST---PNRVA---GYRAPEVTDPRKV 797
Query: 254 NAKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSLVTWATPKLSEDKVRQCVDTR-LGGEY 312
+ K+DVYSFGV+LLELLTG+ P L L W + E+ + D L +
Sbjct: 798 SQKADVYSFGVLLLELLTGKAPTHALLNEEGVDLPRWVQSLVREEWTSEVFDLELLRYQN 977
Query: 313 PPKAVAKMAAVAALCVQYEADFRPNMSIVVKALQPL 348
+ + ++ +A C D RP+++ V ++++ L
Sbjct: 978 VEEEMVQLLQLAVDCAAPYPDRRPSIAEVTRSIEEL 1085
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.315 0.134 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,609,188
Number of Sequences: 28460
Number of extensions: 69040
Number of successful extensions: 760
Number of sequences better than 10.0: 316
Number of HSP's better than 10.0 without gapping: 578
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 582
length of query: 361
length of database: 4,897,600
effective HSP length: 91
effective length of query: 270
effective length of database: 2,307,740
effective search space: 623089800
effective search space used: 623089800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0158.11


