

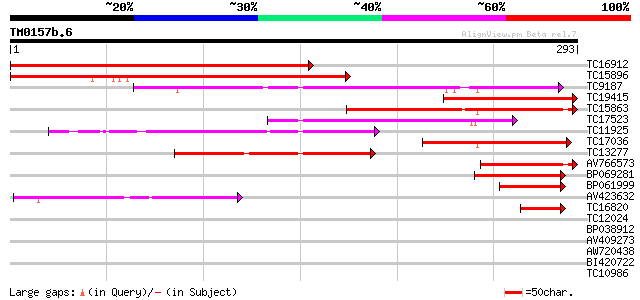
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0157b.6
(293 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC16912 similar to UP|BAD07294 (BAD07294) Prolyl 4-hydroxylase, ... 311 6e-86
TC15896 similar to UP|BAD07294 (BAD07294) Prolyl 4-hydroxylase, ... 238 1e-63
TC9187 similar to UP|Q8LAN3 (Q8LAN3) Prolyl 4-hydroxylase alpha ... 167 1e-42
TC19415 similar to UP|BAD07294 (BAD07294) Prolyl 4-hydroxylase, ... 144 2e-35
TC15863 117 2e-27
TC17523 similar to UP|Q9FKX6 (Q9FKX6) Prolyl 4-hydroxylase, alph... 104 2e-23
TC11925 similar to UP|Q9FKX6 (Q9FKX6) Prolyl 4-hydroxylase, alph... 88 2e-18
TC17036 similar to UP|Q8VZD7 (Q8VZD7) AT3g28480/MFJ20_16, partia... 84 3e-17
TC13277 similar to UP|Q9LSI6 (Q9LSI6) Prolyl 4-hydroxylase alpha... 77 4e-15
AV766573 70 5e-13
BP069281 69 1e-12
BP061999 54 4e-08
AV423632 44 2e-05
TC16820 similar to UP|Q8L8T9 (Q8L8T9) Prolyl 4-hydroxylase alpha... 40 4e-04
TC12024 35 0.011
BP038912 29 1.1
AV409273 28 2.4
AW720438 26 6.9
BI420722 26 6.9
TC10986 similar to GB|AAL56097.1|18028661|AF334030 ORF91 {Helico... 26 9.1
>TC16912 similar to UP|BAD07294 (BAD07294) Prolyl 4-hydroxylase, partial
(30%)
Length = 518
Score = 311 bits (798), Expect = 6e-86
Identities = 157/157 (100%), Positives = 157/157 (100%)
Frame = +1
Query: 1 MKAKTVKGNLSSRSNKLTFPYIFLICIFFFLAGFFGSTLFSHSQDDGRGLRPRPRLLESS 60
MKAKTVKGNLSSRSNKLTFPYIFLICIFFFLAGFFGSTLFSHSQDDGRGLRPRPRLLESS
Sbjct: 46 MKAKTVKGNLSSRSNKLTFPYIFLICIFFFLAGFFGSTLFSHSQDDGRGLRPRPRLLESS 225
Query: 61 EKAEYNLMTAGEFGDDSITSIPFQVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSAL 120
EKAEYNLMTAGEFGDDSITSIPFQVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSAL
Sbjct: 226 EKAEYNLMTAGEFGDDSITSIPFQVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSAL 405
Query: 121 ALREGETEQNTKGIRTSSGVFVSSSEDKTGTLAIIEE 157
ALREGETEQNTKGIRTSSGVFVSSSEDKTGTLAIIEE
Sbjct: 406 ALREGETEQNTKGIRTSSGVFVSSSEDKTGTLAIIEE 516
>TC15896 similar to UP|BAD07294 (BAD07294) Prolyl 4-hydroxylase, partial
(45%)
Length = 613
Score = 238 bits (606), Expect = 1e-63
Identities = 125/185 (67%), Positives = 148/185 (79%), Gaps = 9/185 (4%)
Frame = +3
Query: 1 MKAKTVKGNLSSRSNKLTFPYIFLICIFFFLAGFFGSTLFS--HSQDDGRGLRP--RPR- 55
MK KTV+GN S R+NK+ FP +FL+CIFFFLAGFFGSTLF HSQ+D GLR RPR
Sbjct: 57 MKIKTVRGNWSWRTNKIAFPSVFLLCIFFFLAGFFGSTLFFQYHSQEDEHGLRQIQRPRT 236
Query: 56 --LLES--SEKAEYNLMTAGEFGDDSITSIPFQVLSWKPRALYFPNFATAEQCKSIVGVA 111
LLE+ ++ E +L+ AGE GD+ IT+IPFQVLSW P ALYFPNFATAEQC+SI+ A
Sbjct: 237 TRLLENLTEKETESHLLPAGETGDNFITTIPFQVLSWNPHALYFPNFATAEQCESIIETA 416
Query: 112 KAGLKPSALALREGETEQNTKGIRTSSGVFVSSSEDKTGTLAIIEEKIARATMIPRSHGE 171
K GLKPS L LR GET+++T GIRTSSGVF+S+ EDKTG L +IEEKIARAT IPR+HGE
Sbjct: 417 KEGLKPSTLVLRVGETDESTTGIRTSSGVFISAFEDKTGVLDVIEEKIARATKIPRTHGE 596
Query: 172 AFNIL 176
AFN+L
Sbjct: 597 AFNVL 611
>TC9187 similar to UP|Q8LAN3 (Q8LAN3) Prolyl 4-hydroxylase alpha
subunit-like protein, partial (89%)
Length = 1198
Score = 167 bits (424), Expect = 1e-42
Identities = 101/235 (42%), Positives = 142/235 (59%), Gaps = 13/235 (5%)
Frame = +2
Query: 65 YNLMTAGEFGDDSITSIPFQV--LSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALAL 122
+N +++ G S P +V +SWKPRA + F T +C ++ +AK+ LK SA+A
Sbjct: 110 WNEVSSSYAGSASSIINPSKVKQVSWKPRAFVYEGFLTGLECDHLISLAKSELKRSAVAD 289
Query: 123 R-EGETEQNTKGIRTSSGVFVSSSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVD 181
G+++ + +RTSSG+F+S +D +A IE+KI+ T +P+ +GE +LRYE
Sbjct: 290 NLSGDSKLSE--VRTSSGMFISKKKDPI--VAGIEDKISAWTFLPKENGEDMQVLRYEHG 457
Query: 182 QRYNPHYDSFNPAEYGPQKSQRMASFLLYLTDVQEGGETMFPF------ENGL--NMDVS 233
Q+Y+PHYD F + RMA+ LLYLT+V GGET+FP GL N D+S
Sbjct: 458 QKYDPHYDYFTDKVNIVRGGHRMATVLLYLTNVTRGGETVFPVAEEPPRRRGLETNSDLS 637
Query: 234 YRYEDCI--GLRVRPRQGDGLLFYSLFPNGTIDPTSLHGSCPVIKGEKWVATKWI 286
+C G+ V+PR+GD LLF+SL T DP SLH CPVI+GEKW ATKWI
Sbjct: 638 ----ECAKKGIAVKPRRGDALLFFSLHTTATPDPDSLHAGCPVIEGEKWSATKWI 790
>TC19415 similar to UP|BAD07294 (BAD07294) Prolyl 4-hydroxylase, partial
(24%)
Length = 443
Score = 144 bits (363), Expect = 2e-35
Identities = 65/69 (94%), Positives = 66/69 (95%)
Frame = +3
Query: 225 ENGLNMDVSYRYEDCIGLRVRPRQGDGLLFYSLFPNGTIDPTSLHGSCPVIKGEKWVATK 284
ENGLNMDVSYRYEDC+GLRVRPR GDGLLFYSLFPNGTIDPTSL GSCPVIKGEKWVA K
Sbjct: 3 ENGLNMDVSYRYEDCLGLRVRPRPGDGLLFYSLFPNGTIDPTSLPGSCPVIKGEKWVAPK 182
Query: 285 WIRDHDQDY 293
WIRDHDQDY
Sbjct: 183 WIRDHDQDY 209
>TC15863
Length = 607
Score = 117 bits (294), Expect = 2e-27
Identities = 59/124 (47%), Positives = 81/124 (64%), Gaps = 5/124 (4%)
Frame = +2
Query: 175 ILRYEVDQRYNPHYDSFNPAEYGPQKSQRMASFLLYLTDVQEGGETMFPFENGLNMDVSY 234
+L YEV Q+Y PHYD F QR+A+ L+YL+DV+EGGET+FP V +
Sbjct: 11 VLHYEVGQKYEPHYDYFLDEFNTKNGGQRIATVLMYLSDVEEGGETIFPAAKANFSSVPW 190
Query: 235 RYEDCI-----GLRVRPRQGDGLLFYSLFPNGTIDPTSLHGSCPVIKGEKWVATKWIRDH 289
Y D GL V+P++GD LLF+S+ P+ T+DP+SLHG CPVI+G KW +TKW+ H
Sbjct: 191 -YNDLSVCAKKGLSVKPKRGDALLFWSIRPDATLDPSSLHGGCPVIRGNKWSSTKWM--H 361
Query: 290 DQDY 293
++Y
Sbjct: 362 LEEY 373
>TC17523 similar to UP|Q9FKX6 (Q9FKX6) Prolyl 4-hydroxylase, alpha
subunit-like protein, partial (55%)
Length = 448
Score = 104 bits (260), Expect = 2e-23
Identities = 61/133 (45%), Positives = 76/133 (56%), Gaps = 4/133 (3%)
Frame = +1
Query: 134 IRTSSGVFVSSSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNP 193
+RTSSG F+ DK + IE+KIA T IP HGE +L YEV Q+Y PHYD F
Sbjct: 55 VRTSSGTFLPRGRDKI--VRNIEKKIADFTFIPVEHGEGLQVLHYEVGQKYEPHYDYFLD 228
Query: 194 AEYGPQKSQRMASFLLYLTDVQEGGETMFPFENGLNMDVSYRYE--DC--IGLRVRPRQG 249
QR+A+ L+YLTDV+ GGET+FP G V + E DC GL + P G
Sbjct: 229 EFNTKNGGQRIATVLMYLTDVEXGGETVFPSAKGNFXSVPWWNELSDCGKKGLSINPXXG 408
Query: 250 DGLLFYSLFPNGT 262
D LLF+S+ P T
Sbjct: 409 DALLFWSMNPXAT 447
>TC11925 similar to UP|Q9FKX6 (Q9FKX6) Prolyl 4-hydroxylase, alpha
subunit-like protein, partial (43%)
Length = 613
Score = 87.8 bits (216), Expect = 2e-18
Identities = 57/171 (33%), Positives = 90/171 (52%)
Frame = +1
Query: 21 YIFLICIFFFLAGFFGSTLFSHSQDDGRGLRPRPRLLESSEKAEYNLMTAGEFGDDSITS 80
+IFL+ I L G F ++ D G+ P+P L S + E D+
Sbjct: 121 FIFLVLILLIL----GVLSFPNASSDS-GV-PKPNDLNSIARNAIR----NEGDDNGPRE 270
Query: 81 IPFQVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGV 140
+++SW+PRA + NF T E+C+ ++ +AK ++ S + E ++++ RTSSG
Sbjct: 271 KWVELISWEPRAFLYHNFLTKEECEYLINIAKPHMRKSTVVDSETGKSKDSRA-RTSSGT 447
Query: 141 FVSSSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSF 191
F++ DK + IE++IA T IP HGE +L YEV Q+Y+ HYD F
Sbjct: 448 FLARGRDKI--VRNIEKRIADYTFIPIEHGEGLQVLHYEVGQKYDSHYDYF 594
Score = 25.8 bits (55), Expect = 9.1
Identities = 12/35 (34%), Positives = 19/35 (54%), Gaps = 4/35 (11%)
Frame = -1
Query: 259 PNGTIDPTSLHGSCPVI----KGEKWVATKWIRDH 289
P +I PTSL G C + + E+W +W+ +H
Sbjct: 295 PTKSIQPTSLLGRCHHLHFL*RCERWS*GRWVWEH 191
>TC17036 similar to UP|Q8VZD7 (Q8VZD7) AT3g28480/MFJ20_16, partial (21%)
Length = 697
Score = 84.0 bits (206), Expect = 3e-17
Identities = 43/80 (53%), Positives = 53/80 (65%), Gaps = 3/80 (3%)
Frame = +2
Query: 214 VQEGGETMFPF-ENGLNMDVSYRYEDCI--GLRVRPRQGDGLLFYSLFPNGTIDPTSLHG 270
V +GGET+FP E+ L+ + +C G V+PR+GD LLF+SL N T D SLHG
Sbjct: 2 VGKGGETIFPHAESKLSQPKDESWSECAHKGYAVKPRKGDALLFFSLHLNATTDSNSLHG 181
Query: 271 SCPVIKGEKWVATKWIRDHD 290
SCPVI+GEKW ATKWI D
Sbjct: 182 SCPVIEGEKWSATKWIHVSD 241
>TC13277 similar to UP|Q9LSI6 (Q9LSI6) Prolyl 4-hydroxylase alpha
subunit-like protein, partial (34%)
Length = 582
Score = 76.6 bits (187), Expect = 4e-15
Identities = 42/105 (40%), Positives = 66/105 (62%), Gaps = 1/105 (0%)
Frame = +1
Query: 86 LSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKG-IRTSSGVFVSS 144
LSW PRA F + ++C +V +AK L+ S +A + E+ ++ K +RTSSG+F++
Sbjct: 277 LSWSPRAFLHTGFLSDKECDHLVNLAKDKLEVSMVA--DNESGKSIKSEVRTSSGMFLNK 450
Query: 145 SEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYD 189
++D+ +A IE +IA T +P +GE+ IL YE Q+Y PHYD
Sbjct: 451 AQDEV--VADIEARIATWTFLPIENGESIQILHYENGQKYEPHYD 579
>AV766573
Length = 338
Score = 69.7 bits (169), Expect = 5e-13
Identities = 28/50 (56%), Positives = 41/50 (82%)
Frame = -3
Query: 244 VRPRQGDGLLFYSLFPNGTIDPTSLHGSCPVIKGEKWVATKWIRDHDQDY 293
V+P++GD LLF+S+ P+ T+DP+SLHG CPVI+G KW +TKW+ H ++Y
Sbjct: 336 VKPKRGDALLFWSIRPDATLDPSSLHGGCPVIRGNKWSSTKWM--HLEEY 193
>BP069281
Length = 468
Score = 68.6 bits (166), Expect = 1e-12
Identities = 27/47 (57%), Positives = 37/47 (78%)
Frame = -3
Query: 241 GLRVRPRQGDGLLFYSLFPNGTIDPTSLHGSCPVIKGEKWVATKWIR 287
GL ++P++GD LLF+S+ P+ T D +SLHG CPV KG+KW TKW+R
Sbjct: 421 GLSIKPKRGDALLFWSMKPDTTADTSSLHGGCPVTKGDKWSCTKWMR 281
>BP061999
Length = 564
Score = 53.5 bits (127), Expect = 4e-08
Identities = 21/34 (61%), Positives = 27/34 (78%)
Frame = -2
Query: 254 FYSLFPNGTIDPTSLHGSCPVIKGEKWVATKWIR 287
F+S+ P+ T D +SLHG CPVIKG+KW TKW+R
Sbjct: 563 FWSMKPDTTADTSSLHGGCPVIKGDKWSCTKWMR 462
>AV423632
Length = 484
Score = 44.3 bits (103), Expect = 2e-05
Identities = 33/120 (27%), Positives = 52/120 (42%), Gaps = 2/120 (1%)
Frame = +2
Query: 3 AKTVKGNLSSR--SNKLTFPYIFLICIFFFLAGFFGSTLFSHSQDDGRGLRPRPRLLESS 60
AK L SR S+ T + + F L F + S R P+P L S
Sbjct: 125 AKPRYSRLPSRKSSSSSTLIFALFLAFTFLLLILFALGILSIPSSSSRDKFPKPNDLTS- 301
Query: 61 EKAEYNLMTAGEFGDDSITSIPFQVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSAL 120
+N + + DD +V+SW+PRA + NF T E+C+ ++ +AK + S +
Sbjct: 302 --IAHNTLDRTD-DDDGRGEQWVEVISWEPRAFVYHNFLTKEECEYLIDIAKPNMHKSTV 472
>TC16820 similar to UP|Q8L8T9 (Q8L8T9) Prolyl 4-hydroxylase alpha
subunit-like protein, partial (7%)
Length = 571
Score = 40.4 bits (93), Expect = 4e-04
Identities = 15/23 (65%), Positives = 18/23 (78%)
Frame = +3
Query: 265 PTSLHGSCPVIKGEKWVATKWIR 287
P S+HG C V+ GEKW ATKW+R
Sbjct: 363 PFSVHGGCEVLAGEKWSATKWMR 431
>TC12024
Length = 469
Score = 35.4 bits (80), Expect = 0.011
Identities = 13/37 (35%), Positives = 25/37 (67%)
Frame = +1
Query: 84 QVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSAL 120
++LSW+PRA F NF + +C+ ++ +AK + S++
Sbjct: 334 EILSWEPRAFIFHNFLSKVECEYMINLAKPHMAKSSV 444
>BP038912
Length = 481
Score = 28.9 bits (63), Expect = 1.1
Identities = 10/20 (50%), Positives = 14/20 (70%)
Frame = -2
Query: 266 TSLHGSCPVIKGEKWVATKW 285
+S H CPV+KG+ W A K+
Sbjct: 480 SSYHARCPVLKGDMWSAIKF 421
>AV409273
Length = 417
Score = 27.7 bits (60), Expect = 2.4
Identities = 17/60 (28%), Positives = 27/60 (44%)
Frame = +1
Query: 55 RLLESSEKAEYNLMTAGEFGDDSITSIPFQVLSWKPRALYFPNFATAEQCKSIVGVAKAG 114
R+ + A ++ T GE I S+ + + + PN+A A CKS+V AG
Sbjct: 145 RVFIIASMASEDVKTTGESAVTKIVSLAEEAKLAREGVVKAPNYALASICKSLVAGGVAG 324
>AW720438
Length = 463
Score = 26.2 bits (56), Expect = 6.9
Identities = 8/28 (28%), Positives = 17/28 (60%)
Frame = +3
Query: 86 LSWKPRALYFPNFATAEQCKSIVGVAKA 113
+SW+PR + F + ++C ++ +A A
Sbjct: 357 ISWQPRVFLYKGFLSDKECDYLISLAYA 440
>BI420722
Length = 535
Score = 26.2 bits (56), Expect = 6.9
Identities = 14/36 (38%), Positives = 19/36 (51%), Gaps = 3/36 (8%)
Frame = +2
Query: 83 FQVLSWKPRALYFPNFA---TAEQCKSIVGVAKAGL 115
F VL W P +LYF QC++ VG+ + GL
Sbjct: 215 FWVLCWFPFSLYFAYLVV*YVLYQCRNRVGLVQLGL 322
>TC10986 similar to GB|AAL56097.1|18028661|AF334030 ORF91 {Helicoverpa zea
single nucleocapsid nucleopolyhedrovirus;}, partial
(15%)
Length = 655
Score = 25.8 bits (55), Expect = 9.1
Identities = 9/23 (39%), Positives = 15/23 (65%)
Frame = -2
Query: 19 FPYIFLICIFFFLAGFFGSTLFS 41
FP ++++ +FFFL F +FS
Sbjct: 108 FPILYILLVFFFLLSFSFFVIFS 40
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.319 0.137 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,114,556
Number of Sequences: 28460
Number of extensions: 68344
Number of successful extensions: 399
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 390
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 390
length of query: 293
length of database: 4,897,600
effective HSP length: 90
effective length of query: 203
effective length of database: 2,336,200
effective search space: 474248600
effective search space used: 474248600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0157b.6


