

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0148.17
(215 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
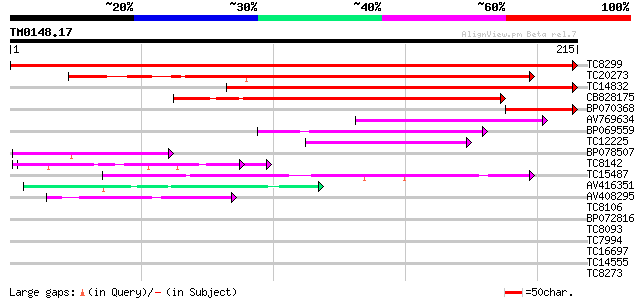
Score E
Sequences producing significant alignments: (bits) Value
TC8299 similar to UP|Q9XEX8 (Q9XEX8) Remorin 1, partial (67%) 422 e-119
TC20273 similar to UP|REMO_SOLTU (P93788) Remorin (pp34), partia... 176 4e-45
TC14832 similar to UP|Q9XEX8 (Q9XEX8) Remorin 1, partial (19%) 145 5e-36
CB828175 113 2e-26
BP070368 57 2e-09
AV769634 47 2e-06
BP069559 45 1e-05
TC12225 44 3e-05
BP078507 43 4e-05
TC8142 UP|BAD11133 (BAD11133) Glutamate-rich protein, complete 42 6e-05
TC15487 similar to UP|O23144 (O23144) Proton pump interactor (AT... 41 1e-04
AV416351 40 2e-04
AV408295 40 3e-04
TC8106 weakly similar to UP|Q9XIV1 (Q9XIV1) mRNA expressed in cu... 38 0.001
BP072816 38 0.002
TC8093 homologue to UP|EFTU_TOBAC (P41342) Elongation factor Tu,... 37 0.002
TC7994 similar to UP|Q8RU73 (Q8RU73) Ribose-5-phosphate isomeras... 37 0.002
TC16697 37 0.003
TC14555 similar to UP|Q43558 (Q43558) Proline rich protein precu... 37 0.003
TC8273 UP|BAD11134 (BAD11134) Glutamate-rich protein, complete 37 0.003
>TC8299 similar to UP|Q9XEX8 (Q9XEX8) Remorin 1, partial (67%)
Length = 1098
Score = 422 bits (1084), Expect = e-119
Identities = 215/215 (100%), Positives = 215/215 (100%)
Frame = +2
Query: 1 MTEEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPS 60
MTEEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPS
Sbjct: 146 MTEEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPS 325
Query: 61 PPESKPVDDSKAIVKVEKTQEAAEEKPLEGSINRDAVLTRVATEKRLSLIKAWEESEKSI 120
PPESKPVDDSKAIVKVEKTQEAAEEKPLEGSINRDAVLTRVATEKRLSLIKAWEESEKSI
Sbjct: 326 PPESKPVDDSKAIVKVEKTQEAAEEKPLEGSINRDAVLTRVATEKRLSLIKAWEESEKSI 505
Query: 121 ADNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEKR 180
ADNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEKR
Sbjct: 506 ADNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEKR 685
Query: 181 AFIEAKKGEDLLKAEELAAKYRATGTAPKKPFSFF 215
AFIEAKKGEDLLKAEELAAKYRATGTAPKKPFSFF
Sbjct: 686 AFIEAKKGEDLLKAEELAAKYRATGTAPKKPFSFF 790
>TC20273 similar to UP|REMO_SOLTU (P93788) Remorin (pp34), partial (64%)
Length = 569
Score = 176 bits (445), Expect = 4e-45
Identities = 101/178 (56%), Positives = 121/178 (67%), Gaps = 1/178 (0%)
Frame = +1
Query: 23 APTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPESKPVDDSKAIVKVEKTQEA 82
A +++TPAP A P P D +K+ PE DDSKA+V V +
Sbjct: 79 AESDTTPAPVAAVPP-------PSNDAVAKKA-------PEVA-ADDSKALVVVPEKTPV 213
Query: 83 AEEKPL-EGSINRDAVLTRVATEKRLSLIKAWEESEKSIADNKAHKKLSDISAWENSKIA 141
E KP +GS++RD L + EKRLS +KAWEESEKS +NKA K LSD+ AWENSK A
Sbjct: 214 PENKPSSKGSLDRDVALAELEKEKRLSYVKAWEESEKSKTENKAQKNLSDVVAWENSKKA 393
Query: 142 AKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEKRAFIEAKKGEDLLKAEELAA 199
A E +LRKIEE LEKKKA Y EK+KNKIA+VH+EAEE+RA IEAK+GEDLLKAEELAA
Sbjct: 394 ALEAQLRKIEERLEKKKAEYGEKMKNKIALVHKEAEERRAMIEAKRGEDLLKAEELAA 567
>TC14832 similar to UP|Q9XEX8 (Q9XEX8) Remorin 1, partial (19%)
Length = 778
Score = 145 bits (366), Expect = 5e-36
Identities = 73/133 (54%), Positives = 99/133 (73%)
Frame = +2
Query: 83 AEEKPLEGSINRDAVLTRVATEKRLSLIKAWEESEKSIADNKAHKKLSDISAWENSKIAA 142
AE + SI+RDAVL RV ++KRL+LIKAWEE+EK+ DNKA+K + WE +K A+
Sbjct: 224 AETDNINTSIDRDAVLARVESQKRLALIKAWEENEKTKVDNKAYKLQCAVDMWEKTKKAS 403
Query: 143 KEVELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEKRAFIEAKKGEDLLKAEELAAKYR 202
+ +++KIEEN+++KKA YVE ++NKIA HR A+EK+A IEA+KGE++LK EE AAK+R
Sbjct: 404 TQAKIKKIEENMDRKKADYVEIMQNKIAETHRLADEKKALIEAQKGEEVLKVEETAAKFR 583
Query: 203 ATGTAPKKPFSFF 215
G PKK S F
Sbjct: 584 TRGYVPKKFLSCF 622
>CB828175
Length = 553
Score = 113 bits (283), Expect = 2e-26
Identities = 58/126 (46%), Positives = 90/126 (71%)
Frame = +2
Query: 63 ESKPVDDSKAIVKVEKTQEAAEEKPLEGSINRDAVLTRVATEKRLSLIKAWEESEKSIAD 122
ES+ + S + + + T++ + P +++DAVL RV ++KRL+LIKAWEE+EK+ D
Sbjct: 125 ESQNAESSNSTLTI--TRQGSTTNP-SYPLDQDAVLARVESQKRLALIKAWEENEKTKVD 295
Query: 123 NKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEKRAF 182
NKA+K + WE +K A+ + +++KIEEN+++KKA YVE ++NKIA HR A+EK+A
Sbjct: 296 NKAYKLQCAVDMWEKTKKASTQAKIKKIEENMDRKKADYVEIMQNKIAETHRLADEKKAL 475
Query: 183 IEAKKG 188
IEAK+G
Sbjct: 476 IEAKRG 493
Score = 32.0 bits (71), Expect = 0.084
Identities = 20/63 (31%), Positives = 35/63 (54%)
Frame = +3
Query: 143 KEVELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEKRAFIEAKKGEDLLKAEELAAKYR 202
K +LRKI ++K + ++ K K+ + ++ ++ K GE++LK EE AAK+R
Sbjct: 366 KSKKLRKI---WTERKLIMLK*CKTKLLKLID*QMRRKH*LKLKGGEEVLKVEETAAKFR 536
Query: 203 ATG 205
G
Sbjct: 537 TRG 545
>BP070368
Length = 300
Score = 57.4 bits (137), Expect = 2e-09
Identities = 27/27 (100%), Positives = 27/27 (100%)
Frame = -3
Query: 189 EDLLKAEELAAKYRATGTAPKKPFSFF 215
EDLLKAEELAAKYRATGTAPKKPFSFF
Sbjct: 298 EDLLKAEELAAKYRATGTAPKKPFSFF 218
>AV769634
Length = 441
Score = 47.4 bits (111), Expect = 2e-06
Identities = 21/73 (28%), Positives = 43/73 (58%)
Frame = -3
Query: 132 ISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMVHREAEEKRAFIEAKKGEDL 191
I+AWEN + A +RK+E LEK++A ++K+ NK+ + +A+E R+ + + + +
Sbjct: 433 ITAWENLQKAKAVAAVRKLEMKLEKRRASSMDKIMNKLRLAKTKAQEMRSSVSPNQTDQV 254
Query: 192 LKAEELAAKYRAT 204
++ A+ + T
Sbjct: 253 VRTSHKASSFLRT 215
>BP069559
Length = 456
Score = 45.1 bits (105), Expect = 1e-05
Identities = 26/87 (29%), Positives = 50/87 (56%)
Frame = -1
Query: 95 DAVLTRVATEKRLSLIKAWEESEKSIADNKAHKKLSDISAWENSKIAAKEVELRKIEENL 154
D L R + E + S + + +E ++ +K H++ + I AWE+ + A E +RK+E L
Sbjct: 453 DEDLQRSSKEAQASSL---DIAESTLDISKFHREEAKIIAWESLQKAKAEAAIRKLEMKL 283
Query: 155 EKKKAVYVEKLKNKIAMVHREAEEKRA 181
EKK++ ++K+ K+ +AE+ R+
Sbjct: 282 EKKRSSSMDKILKKLRRAQMKAEKMRS 202
>TC12225
Length = 552
Score = 43.5 bits (101), Expect = 3e-05
Identities = 21/63 (33%), Positives = 36/63 (56%)
Frame = +3
Query: 113 WEESEKSIADNKAHKKLSDISAWENSKIAAKEVELRKIEENLEKKKAVYVEKLKNKIAMV 172
WEE EK+ + ++ + I AW N + A E + RK+E ++K ++ EKL ++ +V
Sbjct: 117 WEEEEKTKCCLRYQREEAKIQAWINLQNAKAEAQSRKLEVKIQKMRSNLEEKLMKRMTVV 296
Query: 173 HRE 175
H E
Sbjct: 297 HTE 305
>BP078507
Length = 414
Score = 43.1 bits (100), Expect = 4e-05
Identities = 21/62 (33%), Positives = 31/62 (49%), Gaps = 1/62 (1%)
Frame = -1
Query: 2 TEEQPKKVESESPSNPPPPAP-APTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPS 60
T++Q +P + P P+P APT PAP ++ +P P P + + S P PS
Sbjct: 318 TDKQSSPESYPTPLHSPQPSPNAPTSPYPAPSSQLRPPPSTSHQPPPTLLQPTSDTPSPS 139
Query: 61 PP 62
PP
Sbjct: 138 PP 133
Score = 27.3 bits (59), Expect = 2.1
Identities = 22/71 (30%), Positives = 28/71 (38%)
Frame = -1
Query: 2 TEEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSP 61
T QP + S+ P P+P P S+P P P H +P S PSP
Sbjct: 198 TSHQPPPTLLQPTSDTPSPSP-PPPSSPNP-------PPPHHSPNPPHPPSSS----PSP 55
Query: 62 PESKPVDDSKA 72
P+S S A
Sbjct: 54 PQSSQGQRSTA 22
>TC8142 UP|BAD11133 (BAD11133) Glutamate-rich protein, complete
Length = 999
Score = 42.4 bits (98), Expect = 6e-05
Identities = 32/90 (35%), Positives = 46/90 (50%), Gaps = 2/90 (2%)
Frame = +1
Query: 2 TEEQPKKVESES--PSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQP 59
T E+PK+ E+E+ P P P E+ E EAKPE V P+ + EEK+ P+
Sbjct: 280 TTEEPKETETETEAPVAPETQDPLEVENKEVTE-EAKPEEV---KPETEKTEEKTEEPKT 447
Query: 60 SPPESKPVDDSKAIVKVEKTQEAAEEKPLE 89
P + +S A VE+ + AEE P+E
Sbjct: 448 EEPAATTETESAA--PVEEENKPAEEAPVE 531
Score = 40.0 bits (92), Expect = 3e-04
Identities = 32/100 (32%), Positives = 42/100 (42%), Gaps = 4/100 (4%)
Frame = +1
Query: 4 EQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAE-EKSIIPQPSPP 62
EQP E +P P PAP E PA E +P+ + PKE E E + P+ P
Sbjct: 184 EQPAATEVAAPEQPAAEVPAPAE--PATE---EPKEETTEEPKETETETEAPVAPETQDP 348
Query: 63 ---ESKPVDDSKAIVKVEKTQEAAEEKPLEGSINRDAVLT 99
E+K V + +V+ E EEK E A T
Sbjct: 349 LEVENKEVTEEAKPEEVKPETEKTEEKTEEPKTEEPAATT 468
Score = 35.0 bits (79), Expect = 0.010
Identities = 27/89 (30%), Positives = 37/89 (41%), Gaps = 5/89 (5%)
Frame = +1
Query: 3 EEQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPV----VHDAPKEDVAEE-KSIIP 57
E +P +V + P P PA TE APE A P + PKE+ EE K
Sbjct: 133 ENEPTEVTKIQETIPEPEQPAATE-VAAPEQPAAEVPAPAEPATEEPKEETTEEPKETET 309
Query: 58 QPSPPESKPVDDSKAIVKVEKTQEAAEEK 86
+ P + D + E T+EA E+
Sbjct: 310 ETEAPVAPETQDPLEVENKEVTEEAKPEE 396
>TC15487 similar to UP|O23144 (O23144) Proton pump interactor
(AT4g27500/F27G19_100), partial (16%)
Length = 1153
Score = 41.2 bits (95), Expect = 1e-04
Identities = 44/171 (25%), Positives = 75/171 (43%), Gaps = 7/171 (4%)
Frame = +2
Query: 36 KPEPVVHDAPKEDVAEEKSIIPQPSPPESKPVDDSKAIVKVEKTQEAAEEKPLEGSINRD 95
+P+P DA + + ++ P+PSP E+ P ++ KV + ++K LE + +
Sbjct: 206 EPKPAETDAFSKAIPKQPKEEPKPSPQETLPT-QKESKNKVRDLKAKPDKKDLEEADEYE 382
Query: 96 AVLTRVATEKRLSLIKAWEESEKSIADNKAHKKLSDIS----AWENSKIAAKEVELR--- 148
+ R T + E E A K K+ +I+ AWE K A++ +
Sbjct: 383 FEVPRKETPPK--------EPEIDPAKLKEMKREEEIAKAKQAWERKKKLAEKAAAKAAL 538
Query: 149 KIEENLEKKKAVYVEKLKNKIAMVHREAEEKRAFIEAKKGEDLLKAEELAA 199
K ++ EKK +K K K +V A+E +EA + E + E AA
Sbjct: 539 KAQKEAEKKLKDREKKAKKKAGIVEEPADE---VVEAPQEETINDNVEAAA 682
>AV416351
Length = 428
Score = 40.4 bits (93), Expect = 2e-04
Identities = 33/120 (27%), Positives = 48/120 (39%), Gaps = 6/120 (5%)
Frame = +3
Query: 6 PKKVESESPSNPPPPAPAPTESTPAPEAE------AKPEPVVHDAPKEDVAEEKSIIPQP 59
P S P NP PP P P S+ +PE + P P H +PK + + P
Sbjct: 63 PPFSSSSPPRNPRPPPPPPPSSSSSPEKKNSPINHVLPSPTPHQSPK--TIRQSDLSPMN 236
Query: 60 SPPESKPVDDSKAIVKVEKTQEAAEEKPLEGSINRDAVLTRVATEKRLSLIKAWEESEKS 119
S P+SK + + + +Q EE + AVL +K + KAW + S
Sbjct: 237 S-PKSKLSARPPSPLTLPPSQLKTEEPNVPDEAESKAVL----VQKTIDKPKAWHHNNGS 401
>AV408295
Length = 426
Score = 40.0 bits (92), Expect = 3e-04
Identities = 25/72 (34%), Positives = 37/72 (50%)
Frame = +3
Query: 15 SNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPESKPVDDSKAIV 74
S PPPP P+P EAK + V A +E V EE+ + + E KP ++ K
Sbjct: 105 SAPPPP------KAPSPSTEAKKDEVAAAATEEKVEEEEE---KKTEEEKKPAEEEKKEE 257
Query: 75 KVEKTQEAAEEK 86
VE+ ++ AEE+
Sbjct: 258 VVEEEKKPAEEE 293
>TC8106 weakly similar to UP|Q9XIV1 (Q9XIV1) mRNA expressed in cucumber
hypocotyls (Arabinogalactan protein), partial (32%)
Length = 760
Score = 38.1 bits (87), Expect = 0.001
Identities = 27/77 (35%), Positives = 31/77 (40%)
Frame = +1
Query: 14 PSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPESKPVDDSKAI 73
P + PPPAP P S PAP + P P V + P+P SK SK
Sbjct: 118 PVSSPPPAPVPVASPPAPVPVSSPPAPAPTTPAPAVTPTAEV---PAPAPSK----SKKK 276
Query: 74 VKVEKTQEAAEEKPLEG 90
VK K A PL G
Sbjct: 277 VKKGKKHGAPAPSPLLG 327
Score = 32.7 bits (73), Expect = 0.049
Identities = 35/127 (27%), Positives = 47/127 (36%), Gaps = 29/127 (22%)
Frame = +1
Query: 6 PKKVESESPSNPPPPA--PAPTESTPAP----------EAEAKPEPVVHDAPKEDVAEEK 53
P V SP P P + PAP +TPAP A +K + V K
Sbjct: 136 PAPVPVASPPAPVPVSSPPAPAPTTPAPAVTPTAEVPAPAPSKSKKKVKKGKKHGAPAPS 315
Query: 54 SIIPQPSPPESKPVDD---SKAIVKVEKTQEAAEE----KPLEGSI----------NRDA 96
++ P+PP P D S V + AE K + GS+ RD
Sbjct: 316 PLLGPPAPPVGAPGPDDALSPGPTAVAADESGAETIRYLKMVVGSLALGLAVLMF*RRDT 495
Query: 97 VLTRVAT 103
++T VAT
Sbjct: 496 IITAVAT 516
Score = 32.0 bits (71), Expect = 0.084
Identities = 22/76 (28%), Positives = 32/76 (41%)
Frame = +1
Query: 11 SESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPESKPVDDS 70
S++ + P P A AP + PA + P P AP VA + +P SPP P +
Sbjct: 52 SKAAAAPAPVAKAPASTPPAAVPVSSPPP----AP-VPVASPPAPVPVSSPPAPAPTTPA 216
Query: 71 KAIVKVEKTQEAAEEK 86
A+ + A K
Sbjct: 217 PAVTPTAEVPAPAPSK 264
>BP072816
Length = 492
Score = 37.7 bits (86), Expect = 0.002
Identities = 21/60 (35%), Positives = 26/60 (43%), Gaps = 4/60 (6%)
Frame = +1
Query: 11 SESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSII----PQPSPPESKP 66
S SPS+ P AP STP P A +P P A S + P+P+ P S P
Sbjct: 178 STSPSSAPSATAAPPPSTPTPAANTSSKPSTSSKPTTSAAPPASSLLSTPPKPAGPPSNP 357
>TC8093 homologue to UP|EFTU_TOBAC (P41342) Elongation factor Tu,
chloroplast precursor (EF-Tu), partial (58%)
Length = 1146
Score = 37.4 bits (85), Expect = 0.002
Identities = 21/64 (32%), Positives = 28/64 (42%), Gaps = 3/64 (4%)
Frame = +2
Query: 6 PKKVESESPSNPPPP---APAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPP 62
P+ + +NPPPP AP P+ S P + + P+ AP K P PSP
Sbjct: 242 PQSSSTRRRNNPPPPSTAAPPPSPSAPLAASSSARNPMSTSAPSATSTTAKPPSPPPSPW 421
Query: 63 ESKP 66
S P
Sbjct: 422 PSLP 433
>TC7994 similar to UP|Q8RU73 (Q8RU73) Ribose-5-phosphate isomerase
precursor , partial (81%)
Length = 1355
Score = 37.4 bits (85), Expect = 0.002
Identities = 15/49 (30%), Positives = 23/49 (46%)
Frame = +2
Query: 18 PPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPESKP 66
PPP P P+ +P+P+ ++ P + A + P P PP S P
Sbjct: 359 PPPLPNPSVPSPSPKTTSRDSPPTRPSTTSSPAWSSASAPAPPPPSSSP 505
>TC16697
Length = 635
Score = 37.0 bits (84), Expect = 0.003
Identities = 19/61 (31%), Positives = 25/61 (40%), Gaps = 4/61 (6%)
Frame = -1
Query: 14 PSNPPPPA----PAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPESKPVDD 69
P++PPPP+ PAP++ PAP P + S PQP P P
Sbjct: 569 PNHPPPPSSPSPPAPSQRKPAPATAPPSNSSQSPPPNPSSTDPTSPAPQPHPSHLSPASS 390
Query: 70 S 70
S
Sbjct: 389 S 387
>TC14555 similar to UP|Q43558 (Q43558) Proline rich protein precursor,
partial (43%)
Length = 1038
Score = 37.0 bits (84), Expect = 0.003
Identities = 19/51 (37%), Positives = 23/51 (44%)
Frame = +2
Query: 11 SESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSP 61
S P +PPP P P +TP P P P AP + KS P P+P
Sbjct: 422 SPPPFSPPPATPPPPAATPPP--ALTPVPATSPAPAPAKVKSKSPAPAPAP 568
Score = 37.0 bits (84), Expect = 0.003
Identities = 23/81 (28%), Positives = 34/81 (41%), Gaps = 2/81 (2%)
Frame = +2
Query: 9 VESESPSNPPPPAPA-PTESTP-APEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPESKP 66
V ++SPSN P +PA PT +TP P A++ P P P + + P P+ P
Sbjct: 176 VGAQSPSNSPTTSPAPPTPTTPQPPAAQSSPPPAQSSPPPVQSSPPPASTPPPAQSSPPP 355
Query: 67 VDDSKAIVKVEKTQEAAEEKP 87
V + A+ P
Sbjct: 356 VSSPPPVQSTPPPAPASTPPP 418
Score = 33.1 bits (74), Expect = 0.038
Identities = 19/67 (28%), Positives = 27/67 (39%), Gaps = 3/67 (4%)
Frame = +2
Query: 3 EEQPKKVESESP--SNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQP- 59
+ P +S P + PPPA P + +P + P PV P + P P
Sbjct: 257 QSSPPPAQSSPPPVQSSPPPASTPPPAQSSPPPVSSPPPVQSTPPPAPASTPPPASPPPF 436
Query: 60 SPPESKP 66
SPP + P
Sbjct: 437 SPPPATP 457
Score = 33.1 bits (74), Expect = 0.038
Identities = 28/82 (34%), Positives = 34/82 (41%), Gaps = 17/82 (20%)
Frame = +2
Query: 3 EEQPKKVESESPS-NPPPPAPA------------PTESTPAPEAEAKP---EPVVHDAPK 46
+ P V S P + PPPAPA P +TP P A P PV +P
Sbjct: 338 QSSPPPVSSPPPVQSTPPPAPASTPPPASPPPFSPPPATPPPPAATPPPALTPVPATSPA 517
Query: 47 EDVAEEKSIIPQPSP-PESKPV 67
A+ KS P P+P P PV
Sbjct: 518 PAPAKVKSKSPAPAPAPALAPV 583
Score = 32.0 bits (71), Expect = 0.084
Identities = 20/63 (31%), Positives = 27/63 (42%), Gaps = 4/63 (6%)
Frame = +2
Query: 4 EQPKKVESESPSNPPPPAPAP--TESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQP-- 59
+ P + SP+ P P P P +S+P P A++ P PV P P P
Sbjct: 185 QSPSNSPTTSPAPPTPTTPQPPAAQSSP-PPAQSSPPPVQSSPPPASTPPPAQSSPPPVS 361
Query: 60 SPP 62
SPP
Sbjct: 362 SPP 370
Score = 31.6 bits (70), Expect = 0.11
Identities = 21/82 (25%), Positives = 29/82 (34%), Gaps = 2/82 (2%)
Frame = +2
Query: 3 EEQPKKVESESP--SNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPS 60
+ P V+S P S PPP +P + P ++ P P P A P P+
Sbjct: 278 QSSPPPVQSSPPPASTPPPAQSSPPPVSSPPPVQSTPPPAPASTPPP--ASPPPFSPPPA 451
Query: 61 PPESKPVDDSKAIVKVEKTQEA 82
P A+ V T A
Sbjct: 452 TPPPPAATPPPALTPVPATSPA 517
Score = 31.2 bits (69), Expect = 0.14
Identities = 22/72 (30%), Positives = 29/72 (39%), Gaps = 7/72 (9%)
Frame = +2
Query: 2 TEEQPKKVESESPSNPPPPAPAPTESTP-----APEAEAKPEPVVHDAPKEDV--AEEKS 54
T QP +S P P +P P +S+P P A++ P PV P + S
Sbjct: 233 TTPQPPAAQSSPP--PAQSSPPPVQSSPPPASTPPPAQSSPPPVSSPPPVQSTPPPAPAS 406
Query: 55 IIPQPSPPESKP 66
P SPP P
Sbjct: 407 TPPPASPPPFSP 442
Score = 30.8 bits (68), Expect = 0.19
Identities = 20/61 (32%), Positives = 25/61 (40%), Gaps = 7/61 (11%)
Frame = +2
Query: 11 SESPSNPPPPAPAPTES-TPAPEAEAKPEPVV------HDAPKEDVAEEKSIIPQPSPPE 63
S P+ PPPPA P + TP P P P AP +A S+ P +P
Sbjct: 437 SPPPATPPPPAATPPPALTPVPATSPAPAPAKVKSKSPAPAPAPALAPVVSLSPSEAPGP 616
Query: 64 S 64
S
Sbjct: 617 S 619
Score = 25.8 bits (55), Expect = 6.0
Identities = 19/85 (22%), Positives = 28/85 (32%), Gaps = 3/85 (3%)
Frame = +2
Query: 6 PKKVESESPSNPPP---PAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPP 62
P + S+PPP P P + PAP + P +P + P P+
Sbjct: 314 PASTPPPAQSSPPPVSSPPPVQSTPPPAPASTPPPASPPPFSPPPATPPPPAATPPPALT 493
Query: 63 ESKPVDDSKAIVKVEKTQEAAEEKP 87
+ A KV+ A P
Sbjct: 494 PVPATSPAPAPAKVKSKSPAPAPAP 568
>TC8273 UP|BAD11134 (BAD11134) Glutamate-rich protein, complete
Length = 923
Score = 37.0 bits (84), Expect = 0.003
Identities = 24/83 (28%), Positives = 35/83 (41%)
Frame = +1
Query: 4 EQPKKVESESPSNPPPPAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPE 63
E P +VE++ + P TE E K EP+V + + E + + S E
Sbjct: 289 EAPAEVETKEVTEETKPEAEKTEEPEVKPEEVKEEPIVEETKETAATEAAPVEEEKSSEE 468
Query: 64 SKPVDDSKAIVKVEKTQEAAEEK 86
SK + + VE T E A EK
Sbjct: 469 SKAAETT-----VEATTEVAIEK 522
Score = 32.3 bits (72), Expect = 0.064
Identities = 39/137 (28%), Positives = 58/137 (41%), Gaps = 5/137 (3%)
Frame = +1
Query: 20 PAPAPTESTPAPEAEAKPEPVVHDAPKEDVAEEKSIIPQPSPPESKPVDDSKAIV----- 74
PAP PT P+ E EP V E VAE + P E+ ++K +
Sbjct: 175 PAPEPTIEQEHPKEETTEEPKV-----EAVAETTTEAPAAPETEAPAEVETKEVTEETKP 339
Query: 75 KVEKTQEAAEEKPLEGSINRDAVLTRVATEKRLSLIKAWEESEKSIADNKAHKKLSDISA 134
+ EKT+E E KP E + + ++ T++ + A E EKS ++KA
Sbjct: 340 EAEKTEE-PEVKPEE--VKEEPIVEE--TKETAATEAAPVEEEKSSEESKAA-------- 480
Query: 135 WENSKIAAKEVELRKIE 151
E + A EV + K E
Sbjct: 481 -ETTVEATTEVAIEKTE 528
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.302 0.122 0.325
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,344,291
Number of Sequences: 28460
Number of extensions: 67835
Number of successful extensions: 3406
Number of sequences better than 10.0: 602
Number of HSP's better than 10.0 without gapping: 1722
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2708
length of query: 215
length of database: 4,897,600
effective HSP length: 86
effective length of query: 129
effective length of database: 2,450,040
effective search space: 316055160
effective search space used: 316055160
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 17 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.8 bits)
S2: 53 (25.0 bits)
Lotus: description of TM0148.17


