

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0147b.5
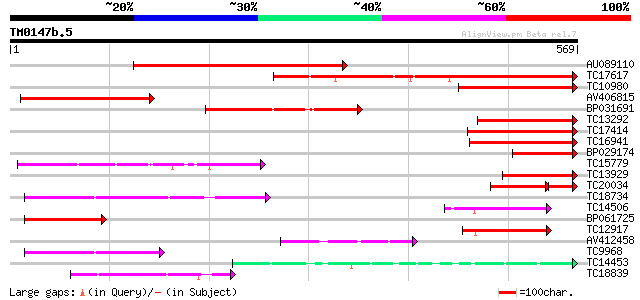
(569 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AU089110 407 e-114
TC17617 weakly similar to UP|Q84SA8 (Q84SA8) Laccase (Fragment),... 258 1e-69
TC10980 similar to UP|Q8VZA1 (Q8VZA1) Laccase (Diphenol oxidase)... 201 2e-52
AV406815 164 3e-41
BP031691 151 3e-37
TC13292 similar to UP|Q9FI28 (Q9FI28) Laccase (Diphenol oxidase)... 147 5e-36
TC17414 weakly similar to UP|Q84SA8 (Q84SA8) Laccase (Fragment),... 144 5e-35
TC16941 similar to UP|Q8L4Y3 (Q8L4Y3) Diphenol oxidase laccase, ... 140 4e-34
BP029174 123 9e-29
TC15779 similar to UP|O24093 (O24093) L-ascorbate oxidase precur... 121 3e-28
TC13929 weakly similar to UP|Q9AUI0 (Q9AUI0) Laccase, partial (14%) 115 1e-26
TC20034 weakly similar to UP|Q9LM34 (Q9LM34) T10O22.11, partial ... 90 3e-26
TC18734 similar to UP|Q8LFM3 (Q8LFM3) Pectinesterase-like protei... 111 4e-25
TC14506 homologue to UP|O24093 (O24093) L-ascorbate oxidase prec... 100 8e-22
BP061725 100 1e-21
TC12917 similar to UP|Q9M5B5 (Q9M5B5) Ascorbate oxidase AO1, par... 93 1e-19
AV412458 92 3e-19
TC9968 similar to UP|Q40473 (Q40473) PS60 protein precursor, par... 86 1e-17
TC14453 similar to PIR|T07634|T07634 pollen-specific protein hom... 83 1e-16
TC18839 similar to GB|BAB01744.1|9280289|AP000603 l-ascorbate ox... 80 9e-16
>AU089110
Length = 646
Score = 407 bits (1046), Expect = e-114
Identities = 199/215 (92%), Positives = 206/215 (95%)
Frame = +1
Query: 125 TLYGPLIILPKHNAQYPFVKPHKEVPIIFGEWWNADTEAVITQALQTGGGPNVSDAYTIN 184
T YGPLIILPKHNAQYPFVKP+KEVP+IFG W+NAD EA+I+QALQTGGGPNVSDAYTIN
Sbjct: 1 TSYGPLIILPKHNAQYPFVKPYKEVPMIFGXWFNADPEAIISQALQTGGGPNVSDAYTIN 180
Query: 185 GLPGPLYNCSQKDTFRLKVKPGKIYLLRLINAALNDELFFSIANHTLTVVEADAGYVKPF 244
GLPGPLYNCS+KDTF+LKVKPG IYLLRLINAALNDELFFSIANHTL VVEADA YVKPF
Sbjct: 181 GLPGPLYNCSKKDTFKLKVKPGXIYLLRLINAALNDELFFSIANHTLKVVEADAVYVKPF 360
Query: 245 VTNTILITPGQTTNVLLKTKSHYPNATFFMTARPYVTGLGTFDNSTVAGILEYKIQHNTT 304
VT TILITPGQTTNVLLKTKSHYPNATFFMTARPYVTGL TFDNSTVAGILEYKIQHN T
Sbjct: 361 VTKTILITPGQTTNVLLKTKSHYPNATFFMTARPYVTGLXTFDNSTVAGILEYKIQHNIT 540
Query: 305 HHNSAVKLKKLPLFKPLLPALNDTSFATKFSNKLR 339
HHNSAVKLKKLPLFKPLLPALNDTSFATKFSNKLR
Sbjct: 541 HHNSAVKLKKLPLFKPLLPALNDTSFATKFSNKLR 645
>TC17617 weakly similar to UP|Q84SA8 (Q84SA8) Laccase (Fragment), partial
(53%)
Length = 1139
Score = 258 bits (660), Expect = 1e-69
Identities = 141/316 (44%), Positives = 192/316 (60%), Gaps = 11/316 (3%)
Frame = +2
Query: 265 SHYPNATFFMTARPYVTGLGTFDNSTVAGILEYKIQHNTTHHNSAVKLKKLPLFKPLLPA 324
++ P +++M A PY +G FDN T GI+ Y Q+ + +S + + LP P++P
Sbjct: 20 ANQPIGSYYMVASPYKSGGKMFDNITSRGIVVYDHQY-AQYSSSHLVMPSLPSLNPMMPF 196
Query: 325 L---NDTSFATKFSNKLRSLASARFPANVPQKVDKHFLFTVGLGTNPCK--NKSNQTCQG 379
L NDT A KF + + SL A VP +VD+H TVGL C +N TC+G
Sbjct: 197 LPPFNDTPTAHKFYSNITSLVGAPHWVPVPLEVDEHMFITVGLNLQRCDPATVTNDTCKG 376
Query: 380 PNNAAMFSASVNNVSFILPTT---ALLQAHFFGKSNGVYTPDFPTSPLHPFNYTGTTTPN 436
P N + S S+NN SF+LP ++L+A FF +GVYT DFP+ P F++T
Sbjct: 377 PMNHTL-SPSMNNESFVLPKGRGYSMLEA-FFENMSGVYTTDFPSKPPVMFDFTNPDIRF 550
Query: 437 NTNV---SNGTKVVVLPFNTSVELVMQDTSILGAESHPLHLHGFNFFVVGQGFGNFDPNK 493
+ N+ TKV L FN++VE+V Q+T+ + ++H +HLHGFNF V+ QGFGNFDP
Sbjct: 551 DLNLIFAPKSTKVKKLKFNSTVEIVFQNTAFIHTDNHAMHLHGFNFHVLAQGFGNFDPTS 730
Query: 494 DPSKFNLVDPVERNTVGVPSGGWVAIRFLADNPGVWFMHCHLEVHTSWGLRMAWVVLDGK 553
D +KFNLV+P RNT+ VP GGW IRF A+NPGVWF+HCH+E HT G MA+ V +G
Sbjct: 731 DEAKFNLVNPQIRNTIAVPVGGWAVIRFQANNPGVWFVHCHVEHHTHRGFNMAFEVENGP 910
Query: 554 QPNQKLLPPPTDLPKC 569
+ L PPP DLPKC
Sbjct: 911 TLSTSLTPPPADLPKC 958
>TC10980 similar to UP|Q8VZA1 (Q8VZA1) Laccase (Diphenol oxidase)-like
protein, partial (22%)
Length = 572
Score = 201 bits (512), Expect = 2e-52
Identities = 87/119 (73%), Positives = 102/119 (85%)
Frame = +1
Query: 451 FNTSVELVMQDTSILGAESHPLHLHGFNFFVVGQGFGNFDPNKDPSKFNLVDPVERNTVG 510
FN++VELV+QDT++L ESHP HLHG+NFFVVG G GNFDP+KDP+K+NLVDPVERNTVG
Sbjct: 7 FNSTVELVLQDTNLLSVESHPFHLHGYNFFVVGTGVGNFDPSKDPAKYNLVDPVERNTVG 186
Query: 511 VPSGGWVAIRFLADNPGVWFMHCHLEVHTSWGLRMAWVVLDGKQPNQKLLPPPTDLPKC 569
VP+GGW AIRF ADNPGVWF HCHLE+HT WGL+ A+VV DG +Q +LPPP DLP C
Sbjct: 187 VPTGGWTAIRFRADNPGVWFFHCHLELHTGWGLKTAFVVEDGPGQDQSVLPPPKDLPAC 363
>AV406815
Length = 426
Score = 164 bits (416), Expect = 3e-41
Identities = 74/134 (55%), Positives = 95/134 (70%)
Frame = +2
Query: 12 AGTTRHYQFDIRYQNVTRLCHSKSLVTVNGQFPGPRIIAREGDRLLIKVVNHVQNNISIH 71
+GT ++F I+ V RLC ++ ++TVNGQFPGP + R GD + IKV N NISIH
Sbjct: 23 SGTHHSHEFVIQTTPVKRLCRTQRILTVNGQFPGPTVQVRNGDSVAIKVTNAGPYNISIH 202
Query: 72 WHGIRQLQSGWADGPAYVTQCPIQTGQSYVYNYTIKGQRGTLFWHAHISWLRATLYGPLI 131
WHG+R L++ WADGP+YVTQCPIQ G SY Y +TI+ Q GTL+WHAH +LRAT+YG I
Sbjct: 203 WHGLRMLRNPWADGPSYVTQCPIQPGGSYTYRFTIQNQEGTLWWHAHTGFLRATVYGAFI 382
Query: 132 ILPKHNAQYPFVKP 145
I P+ + YPF P
Sbjct: 383 IYPRLGSPYPFSMP 424
>BP031691
Length = 481
Score = 151 bits (381), Expect = 3e-37
Identities = 85/162 (52%), Positives = 109/162 (66%), Gaps = 4/162 (2%)
Frame = +1
Query: 197 DTFRLKVKPGKIYLLRLINAALNDELFFSIANHTLTVVEADAGYVKPFVTNTILITPGQT 256
DTF V+ GK YLLR+INAAL+DEL+F +ANHTLTVVE DA Y KPF T I++TPGQT
Sbjct: 13 DTFIYTVESGKTYLLRIINAALSDELYFGVANHTLTVVEVDASYTKPFNTRAIMVTPGQT 192
Query: 257 TNVLLKTKSHYPNAT--FFMTARPYVTGLGTFDNSTVAGILEYKIQHNTTHHNSAVKLKK 314
TNVLL+ + P+++ F M ARPY T + FDNST G L Y N+T VK
Sbjct: 193 TNVLLRA-NQIPDSSGLFVMAARPYRTSVFPFDNSTTIGYLRY----NST-RGEKVKPPH 354
Query: 315 LPLFKPL--LPALNDTSFATKFSNKLRSLASARFPANVPQKV 354
+P + LP + DT F T+FS KLRS+A+++FP VP+K+
Sbjct: 355 VPTDLSIHNLPEMEDTKFETEFSAKLRSIATSQFPCKVPKKI 480
>TC13292 similar to UP|Q9FI28 (Q9FI28) Laccase (Diphenol oxidase), partial
(19%)
Length = 558
Score = 147 bits (371), Expect = 5e-36
Identities = 62/100 (62%), Positives = 75/100 (75%)
Frame = +2
Query: 470 HPLHLHGFNFFVVGQGFGNFDPNKDPSKFNLVDPVERNTVGVPSGGWVAIRFLADNPGVW 529
HP+HLHGF+F VVG GFGNF+ K P ++L+DP NTV VP GW AIRF+ADNPGVW
Sbjct: 8 HPMHLHGFSFHVVGYGFGNFNKRKSPMYYDLIDPPLLNTVIVPKNGWAAIRFVADNPGVW 187
Query: 530 FMHCHLEVHTSWGLRMAWVVLDGKQPNQKLLPPPTDLPKC 569
FMHCHLE H SWG+ ++V +GK + KLLPPP D+P C
Sbjct: 188 FMHCHLERHLSWGMETVFIVKNGKSSHAKLLPPPPDMPPC 307
>TC17414 weakly similar to UP|Q84SA8 (Q84SA8) Laccase (Fragment), partial
(27%)
Length = 670
Score = 144 bits (362), Expect = 5e-35
Identities = 60/110 (54%), Positives = 81/110 (73%)
Frame = +3
Query: 460 QDTSILGAESHPLHLHGFNFFVVGQGFGNFDPNKDPSKFNLVDPVERNTVGVPSGGWVAI 519
Q+T++L A+SHP+H+HG+NF V+ Q FG ++P D +K+NLV+P TV VP GGW AI
Sbjct: 15 QNTAVLNAQSHPMHIHGYNFHVLAQDFGIYNPILDKAKYNLVNPQISTTVPVPPGGWAAI 194
Query: 520 RFLADNPGVWFMHCHLEVHTSWGLRMAWVVLDGKQPNQKLLPPPTDLPKC 569
RF A+NPG WF+HCH++ H WGL ++V +G P+ LLPPP DLPKC
Sbjct: 195 RFQANNPGAWFVHCHVDDHNVWGLTTVFLVENGPTPSTSLLPPPADLPKC 344
>TC16941 similar to UP|Q8L4Y3 (Q8L4Y3) Diphenol oxidase laccase, partial
(18%)
Length = 625
Score = 140 bits (354), Expect = 4e-34
Identities = 61/108 (56%), Positives = 77/108 (70%)
Frame = +2
Query: 462 TSILGAESHPLHLHGFNFFVVGQGFGNFDPNKDPSKFNLVDPVERNTVGVPSGGWVAIRF 521
TSI+ E+HP+HLHG++F+VV +GFGNF+P K SKFNL+DP R TV VP+ GW IRF
Sbjct: 2 TSIVTPENHPIHLHGYDFYVVAEGFGNFNPKKHTSKFNLLDPPMRYTVSVPANGWAVIRF 181
Query: 522 LADNPGVWFMHCHLEVHTSWGLRMAWVVLDGKQPNQKLLPPPTDLPKC 569
+ADNPG W MHCHL+VH WGL +V +G + + PP DLP C
Sbjct: 182 VADNPGAWIMHCHLDVHIGWGLATILLVDNGVGLLESIESPPEDLPLC 325
>BP029174
Length = 493
Score = 123 bits (308), Expect = 9e-29
Identities = 51/65 (78%), Positives = 59/65 (90%)
Frame = -2
Query: 505 ERNTVGVPSGGWVAIRFLADNPGVWFMHCHLEVHTSWGLRMAWVVLDGKQPNQKLLPPPT 564
ERNTVGVP+GGW AIRF ADNPGVWFMHCHLE+HT+WGL+MA+VV +GK PN+ +LPPP
Sbjct: 492 ERNTVGVPAGGWTAIRFRADNPGVWFMHCHLEIHTTWGLKMAFVVDNGKGPNESILPPPR 313
Query: 565 DLPKC 569
DLPKC
Sbjct: 312 DLPKC 298
>TC15779 similar to UP|O24093 (O24093) L-ascorbate oxidase precursor,
partial (50%)
Length = 1304
Score = 121 bits (304), Expect = 3e-28
Identities = 92/276 (33%), Positives = 135/276 (48%), Gaps = 28/276 (10%)
Frame = +1
Query: 9 LALAGTTRHYQFDIRYQNVTRLCHSKSLVTVNGQFPGPRIIAREGDRLLIKVVN--HVQN 66
+ + RHY+FD+ Y ++ +NGQFPGP I A GD L I + N H +
Sbjct: 175 VVVGAKVRHYKFDVEYMFKKPDGLEHVVMGINGQFPGPTIKAEVGDTLDIALTNKLHTEG 354
Query: 67 NISIHWHGIRQLQSGWADGPAYVTQCPIQTGQSYVYNYTIKGQRGTLFWHAHISWLRAT- 125
+ IH HGIRQL + WADG A ++QC I G+++ Y +T+ + GT F+H H RA
Sbjct: 355 TV-IHGHGIRQLGTPWADGTAAISQCAINPGETFHYWFTV-DRPGTYFYHGHYGMQRAAG 528
Query: 126 LYGPLII-LPKHNAQYPFVKPHKEVPIIFGEWWNADTE----AVITQALQTGGGPNVSDA 180
LYG LI+ LPK + PF E ++ + W+ + + ++ L+ G P +
Sbjct: 529 LYGSLIVDLPKGQNE-PF-HYDGEFNLLLSDLWHKSSHEQEVGLSSKPLKWIGEP---QS 693
Query: 181 YTINGLPGPLYNCSQKDTF--------------------RLKVKPGKIYLLRLINAALND 220
ING +NCS F L V+P K Y +R+ +
Sbjct: 694 LLING--RGQFNCSLAAKFVNTSLVPQCQFKGGEECAPEILHVEPNKTYRIRIASTTSMA 867
Query: 221 ELFFSIANHTLTVVEADAGYVKPFVTNTILITPGQT 256
L +I+NH L VVEAD +V+PF + I I G+T
Sbjct: 868 SLNLAISNHKLIVVEADGNHVQPFAVDDIDIYSGET 975
>TC13929 weakly similar to UP|Q9AUI0 (Q9AUI0) Laccase, partial (14%)
Length = 484
Score = 115 bits (289), Expect = 1e-26
Identities = 45/75 (60%), Positives = 61/75 (81%)
Frame = +1
Query: 495 PSKFNLVDPVERNTVGVPSGGWVAIRFLADNPGVWFMHCHLEVHTSWGLRMAWVVLDGKQ 554
P++FNLVDP +NT+G P GGWVAIRF+ADNPG+WF+HCH++ H +WGL A +V +G
Sbjct: 1 PARFNLVDPPVKNTIGTPPGGWVAIRFVADNPGIWFLHCHIDSHLNWGLATALLVENGVG 180
Query: 555 PNQKLLPPPTDLPKC 569
P+Q ++PPP DLP+C
Sbjct: 181 PSQSVIPPPPDLPQC 225
>TC20034 weakly similar to UP|Q9LM34 (Q9LM34) T10O22.11, partial (15%)
Length = 457
Score = 90.1 bits (222), Expect(2) = 3e-26
Identities = 38/60 (63%), Positives = 43/60 (71%)
Frame = +2
Query: 483 GQGFGNFDPNKDPSKFNLVDPVERNTVGVPSGGWVAIRFLADNPGVWFMHCHLEVHTSWG 542
G GFGNFD KDP +NLVDP +NTV +P GW IRF A NPGVWFMHCH+E H + G
Sbjct: 2 GWGFGNFD*EKDPLGYNLVDPPYQNTVAIPKNGWTTIRFRAKNPGVWFMHCHIERHVTLG 181
Score = 45.4 bits (106), Expect(2) = 3e-26
Identities = 15/29 (51%), Positives = 22/29 (75%)
Frame = +3
Query: 541 WGLRMAWVVLDGKQPNQKLLPPPTDLPKC 569
WG+ M ++V DG P +K+LPPP D+P+C
Sbjct: 177 WGMAMTFIVKDGNNPEEKMLPPPLDMPQC 263
>TC18734 similar to UP|Q8LFM3 (Q8LFM3) Pectinesterase-like protein, partial
(47%)
Length = 929
Score = 111 bits (277), Expect = 4e-25
Identities = 63/247 (25%), Positives = 118/247 (47%), Gaps = 1/247 (0%)
Frame = +3
Query: 16 RHYQFDIRYQNVTRLCHSKSLVTVNGQFPGPRIIAREGDRLLIKVVNHVQNNISIHWHGI 75
+ Y + + Y ++ L + ++ +NGQFPGPR+ D +++ ++N + + W+GI
Sbjct: 84 KFYTWTVTYGTLSPLGSPQQVILINGQFPGPRLDLVTNDNVILNLINKLDEPFLLTWNGI 263
Query: 76 RQLQSGWADGPAYVTQCPIQTGQSYVYNYTIKGQRGTLFWHAHISWLRAT-LYGPLIILP 134
+Q ++ W DG T CPI +Y Y + K Q GT + +A+ +G L +L
Sbjct: 264 KQRKNSWQDG-VLGTNCPIPPNSNYTYKFQAKDQIGTYTYFPSTKMHKASGGFGALNVLH 440
Query: 135 KHNAQYPFVKPHKEVPIIFGEWWNADTEAVITQALQTGGGPNVSDAYTINGLPGPLYNCS 194
+ P+ P + ++ G+W+ + ++Q L +G D ING + +
Sbjct: 441 RSVIPIPYPNPDGDFTLLIGDWYKT-SHKTLSQTLDSGKSLAFPDGLLINGQAHSTFTVN 617
Query: 195 QKDTFRLKVKPGKIYLLRLINAALNDELFFSIANHTLTVVEADAGYVKPFVTNTILITPG 254
Q GK Y+ R+ N L+ + F I HTL +VE + + +++ + G
Sbjct: 618 Q----------GKTYMFRISNVGLSTSINFRIQGHTLKLVEVEGSHTIQNEYDSLDVHVG 767
Query: 255 QTTNVLL 261
Q+ +VL+
Sbjct: 768 QSVSVLV 788
>TC14506 homologue to UP|O24093 (O24093) L-ascorbate oxidase precursor,
partial (24%)
Length = 692
Score = 100 bits (248), Expect = 8e-22
Identities = 50/110 (45%), Positives = 65/110 (58%), Gaps = 3/110 (2%)
Frame = +3
Query: 437 NTNVSNGTKVVVLPFNTSVELVMQDTSIL---GAESHPLHLHGFNFFVVGQGFGNFDPNK 493
NT V NG V + N V++++Q+ + L G+E HP HLHG +F+V+G G G F P
Sbjct: 30 NTTVGNG--VYMFQLNEVVDVILQNANQLNGNGSEIHPWHLHGHDFWVLGYGEGKFRPGV 203
Query: 494 DPSKFNLVDPVERNTVGVPSGGWVAIRFLADNPGVWFMHCHLEVHTSWGL 543
D FNL RNT + GW A+RF ADNPGVW HCH+E H G+
Sbjct: 204 DEKSFNLTRAPLRNTAVIFPYGWTALRFKADNPGVWAFHCHIEPHLHMGM 353
>BP061725
Length = 358
Score = 99.8 bits (247), Expect = 1e-21
Identities = 40/82 (48%), Positives = 60/82 (72%)
Frame = +3
Query: 16 RHYQFDIRYQNVTRLCHSKSLVTVNGQFPGPRIIAREGDRLLIKVVNHVQNNISIHWHGI 75
RH++F++R + T+LC +K+++TVNGQFPGP + +G+ +++ V N NI+IH HG+
Sbjct: 108 RHHKFEVREVSYTKLCSTKNILTVNGQFPGPTLYVTKGETIIVHVYNRANYNITIHXHGV 287
Query: 76 RQLQSGWADGPAYVTQCPIQTG 97
Q + W+DGP YVTQCPIQ G
Sbjct: 288 NQPRYPWSDGPEYVTQCPIQPG 353
>TC12917 similar to UP|Q9M5B5 (Q9M5B5) Ascorbate oxidase AO1, partial (19%)
Length = 521
Score = 93.2 bits (230), Expect = 1e-19
Identities = 43/93 (46%), Positives = 61/93 (65%), Gaps = 4/93 (4%)
Frame = +1
Query: 455 VELVMQDTSILG----AESHPLHLHGFNFFVVGQGFGNFDPNKDPSKFNLVDPVERNTVG 510
V++V+Q+T+IL ++ HP HLHG +F+++G G G F P D +KFNL +P RN
Sbjct: 1 VDVVLQNTNILERIKRSDFHPWHLHGHDFWILGYGDGMFKPGDDETKFNLENPPLRNNAV 180
Query: 511 VPSGGWVAIRFLADNPGVWFMHCHLEVHTSWGL 543
+ GW A+RF A+NPGVW HCH+E H G+
Sbjct: 181 LFPYGWTALRFKANNPGVWAFHCHVEPHLHMGM 279
>AV412458
Length = 423
Score = 91.7 bits (226), Expect = 3e-19
Identities = 56/140 (40%), Positives = 81/140 (57%), Gaps = 2/140 (1%)
Frame = +2
Query: 272 FFMTARPYVTGLGT-FDNSTVAGILEYKIQHNTTHHNSAVKLKKLPLFKPLLPALNDTSF 330
++M AR Y T + FDN+T ILEY ++ ++ + PLLPA NDT
Sbjct: 23 YYMAARAYQTAMNAAFDNTTTTAILEYIPRNRNVQPSTPIL--------PLLPAFNDTPT 178
Query: 331 ATKFSNKLRSLASARFPANVPQKVDKHFLFTVGLGTNPCKNKSNQTCQGPNNAAMFSASV 390
AT F++ +R L+ N D + FTVGLG C+N ++ CQGP N + F+AS+
Sbjct: 179 ATAFTSGIRGLSQNVVFKN---SADVNLFFTVGLGLIFCRNPNSPNCQGP-NGSRFAASI 346
Query: 391 NNVSFILP-TTALLQAHFFG 409
NN SF+LP TT+L+QA++ G
Sbjct: 347 NNNSFVLPRTTSLMQAYYQG 406
>TC9968 similar to UP|Q40473 (Q40473) PS60 protein precursor, partial (27%)
Length = 631
Score = 86.3 bits (212), Expect = 1e-17
Identities = 43/141 (30%), Positives = 78/141 (54%), Gaps = 1/141 (0%)
Frame = +2
Query: 16 RHYQFDIRYQNVTRLCHSKSLVTVNGQFPGPRIIAREGDRLLIKVVNHVQNNISIHWHGI 75
R + + I Y ++ L + + +NGQFPGP I + D L+I V NH+ + W+G+
Sbjct: 212 RFFDWTITYGDIYPLGVKQQGILINGQFPGPEIYSVTNDNLVINVHNHLPEPFLLSWNGV 391
Query: 76 RQLQSGWADGPAYVTQCPIQTGQSYVYNYTIKGQRGTLFWHAHISWLRAT-LYGPLIILP 134
+Q ++ + DG Y T CPI G+++ YN +K Q G+ F+ +++ +A +G + IL
Sbjct: 392 QQRRNSYQDG-VYGTTCPIPPGKNFTYNLQVKDQIGSFFYFPSLAFHKAAGGFGAIKILS 568
Query: 135 KHNAQYPFVKPHKEVPIIFGE 155
+ PF +P + ++ G+
Sbjct: 569 RPRIPVPFPEPAADFSLLIGD 631
>TC14453 similar to PIR|T07634|T07634 pollen-specific protein homolog
T1P17.10 - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (56%)
Length = 1364
Score = 83.2 bits (204), Expect = 1e-16
Identities = 95/352 (26%), Positives = 143/352 (39%), Gaps = 6/352 (1%)
Frame = +2
Query: 224 FSIANHTLTVVEADAGYVKPFVTNTILITPGQTTNVLLKTKSHYPNATFFMTARPYVTGL 283
F I NH L + E + Y ++ + GQ+ L+ T + +++ A
Sbjct: 2 FRIQNHNLLLAETEGSYTVQQNYTSLDVHVGQSYTFLV-TMDQNASTDYYIVASARFVNE 178
Query: 284 GTFDNSTVAGILEYKIQHNTTHHNSAVKLKKLPLFKPLLPALNDTSFATKFSNKLRSL-- 341
+ T GIL Y NS K + PL A D T N+ RS+
Sbjct: 179 SRWQRVTGVGILHYT--------NSKGKARG-----PLPAAPEDQFDKTYSMNQARSIRW 319
Query: 342 -ASARFPANVPQKVDKHFLFTVGLGTNPCKNKSNQTCQGPNNAAMFSASVNNVSFILPTT 400
SA PQ ++ V KNK G A + + +SF+ P+T
Sbjct: 320 NVSASGARPNPQGSFRYGSINV-TEVYVLKNKPRVKINGKKRATL-----SGISFVNPST 481
Query: 401 ALLQAHFFGKSNGVYTPDFPTSPLHPFNYTGTTTPN-NTNVSNGTKVVVLPFNTSVELVM 459
+ A F K +Y DFPT PL T +P T+V NG+ + +E+++
Sbjct: 482 PIRLADQF-KLKDIYKLDFPTRPL-------TGSPRAETSVINGS------YRGFIEIIL 619
Query: 460 QDTSILGAESHPLHLHGFNFFVVGQGFGNFDPNKDPSKFNLVDPVERNTVGVPSGGWVAI 519
Q+ H HL G+ FFVVG +G++ N + +N D + R T V G W AI
Sbjct: 620 QNND---TRMHTYHLDGYAFFVVGMDYGDWSDNSRGT-YNKWDGIARATTQVYPGAWTAI 787
Query: 520 RFLADNPGVWFMHCHLEVHTSWGL-RMAWVVLDGKQPNQKL-LPPPTDLPKC 569
DN G+W + E SW L + ++ + +PN K LP P + C
Sbjct: 788 LVSLDNVGIW--NLRTENLDSWYLGQETYIRVVNPEPNNKTELPIPDNALFC 937
>TC18839 similar to GB|BAB01744.1|9280289|AP000603 l-ascorbate oxidase;
pectinesterase-like protein; pollen-specific
protein-like {Arabidopsis thaliana;} , partial (29%)
Length = 498
Score = 80.1 bits (196), Expect = 9e-16
Identities = 53/173 (30%), Positives = 79/173 (45%), Gaps = 8/173 (4%)
Frame = +1
Query: 62 NHVQNNISIHWHGIRQLQSGWADGPAYVTQCPIQTGQSYVYNYTIKGQRGTLFWHAHISW 121
N++ + WHG++Q ++ W DG A T CPI G +Y Y++ +K Q G+ F++ +
Sbjct: 13 NNLDEPLQFTWHGVQQRKNSWQDGVAG-TSCPILPGTNYTYHFQVKDQIGSYFYYPTTAL 189
Query: 122 LRAT-LYGPLIILPKHNAQYPFVKPHKEVPIIFGEWWNADTEAVITQALQTGGGPNVSDA 180
RA +G L I + P+ P E +I G+W+ A + + L +G D
Sbjct: 190 HRAAGGFGGLRIFSRLLIPVPYPDPEDEYWVIIGDWY-AKSHKTLKSFLDSGRSIGRPDG 366
Query: 181 YTINGLPG-------PLYNCSQKDTFRLKVKPGKIYLLRLINAALNDELFFSI 226
ING PLY +KPGK Y R+ N L D L F I
Sbjct: 367 VLINGKTAKGDGSDQPLYT----------MKPGKTYKYRICNVGLKDSLNFRI 495
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.137 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,397,954
Number of Sequences: 28460
Number of extensions: 174573
Number of successful extensions: 967
Number of sequences better than 10.0: 69
Number of HSP's better than 10.0 without gapping: 943
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 951
length of query: 569
length of database: 4,897,600
effective HSP length: 95
effective length of query: 474
effective length of database: 2,193,900
effective search space: 1039908600
effective search space used: 1039908600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0147b.5


