

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0147b.4
(313 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
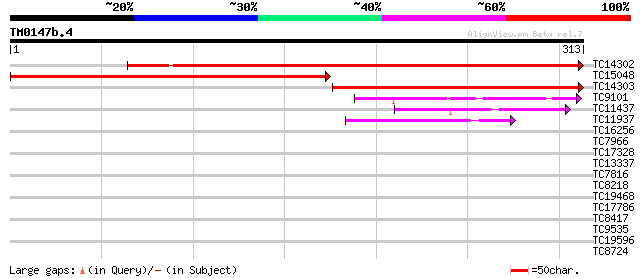
Score E
Sequences producing significant alignments: (bits) Value
TC14302 similar to UP|Y230_ARATH (O80934) Protein At2g37660, chl... 429 e-121
TC15048 similar to UP|Y230_ARATH (O80934) Protein At2g37660, chl... 346 3e-96
TC14303 similar to UP|Y230_ARATH (O80934) Protein At2g37660, chl... 270 2e-73
TC9101 similar to GB|AAL08281.1|15912295|AY056425 At2g34460/T31E... 58 2e-09
TC11437 homologue to GB|AAP37706.1|30725368|BT008347 At4g31530 {... 56 9e-09
TC11937 weakly similar to UP|AAS49102 (AAS49102) At1g72640, part... 52 1e-07
TC16256 similar to UP|Q7Y0H8 (Q7Y0H8) Cinnamoyl CoA reductase, p... 39 0.001
TC7966 similar to UP|Q84V25 (Q84V25) Quinone oxidoreductase, par... 38 0.002
TC17328 weakly similar to UP|Q9FGH3 (Q9FGH3) Dihydroflavonol 4-r... 28 1.5
TC13337 28 2.6
TC7816 homologue to UP|Q9ATM4 (Q9ATM4) Plasma membrane integral ... 27 4.5
TC8218 homologue to UP|Q93VR3 (Q93VR3) AT5g28840/F7P1_20, complete 27 5.8
TC19468 similar to UP|Q7VB79 (Q7VB79) Predicted protein family P... 27 5.8
TC17786 similar to PIR|T46177|T46177 villin 3 homolog T8H10.10 -... 27 5.8
TC8417 similar to UP|GAE1_PEA (Q43070) UDP-glucose 4-epimerase ... 27 5.8
TC9535 26 7.6
TC19596 26 7.6
TC8724 homologue to UP|Q9AQU5 (Q9AQU5) Plasma membrane integral ... 26 9.9
>TC14302 similar to UP|Y230_ARATH (O80934) Protein At2g37660, chloroplast
precursor, partial (75%)
Length = 1066
Score = 429 bits (1103), Expect = e-121
Identities = 211/249 (84%), Positives = 233/249 (92%)
Frame = +2
Query: 65 STVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTIGASDDVYVGDIRDTGSIA 124
STVLVTGAGGRTG+IVYK L+ER QY+ARGLVRTEESKQ IG +DDV++GDIRD GSI
Sbjct: 68 STVLVTGAGGRTGQIVYKTLKER--QYVARGLVRTEESKQKIGGADDVFIGDIRDAGSIV 241
Query: 125 PAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKNQIDAAKAAG 184
PAIQGIDALIILTSA P MKPGF+PTKG RPEFYF+DGAYPEQVDWIGQKNQIDAAKAAG
Sbjct: 242 PAIQGIDALIILTSATPKMKPGFDPTKGGRPEFYFDDGAYPEQVDWIGQKNQIDAAKAAG 421
Query: 185 VKQIVLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGGLQDKEGGV 244
VK++VLVGSMGGT+LN+PLNSLGNGNILVWKRKAEQYLADSGIPYTIIR GGL DK+GG+
Sbjct: 422 VKRVVLVGSMGGTNLNHPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRPGGLLDKDGGL 601
Query: 245 RELIIGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDLASKPEGAGTPTRDFKA 304
RELI+GKDDE+L+TET+TI R DVAEVC+QALN+EE QFKAFDLASKPEG GTPT+DFKA
Sbjct: 602 RELILGKDDELLQTETKTIPRADVAEVCVQALNYEETQFKAFDLASKPEGVGTPTKDFKA 781
Query: 305 LFSQITTRF 313
LFSQIT+RF
Sbjct: 782 LFSQITSRF 808
>TC15048 similar to UP|Y230_ARATH (O80934) Protein At2g37660, chloroplast
precursor, partial (34%)
Length = 692
Score = 346 bits (888), Expect = 3e-96
Identities = 175/175 (100%), Positives = 175/175 (100%)
Frame = +3
Query: 1 MGMATRVPFVSATLSPNQCHKYCLPSSLKLLTSLQFSHSSSLSLATHKRVRTRTSVVAMA 60
MGMATRVPFVSATLSPNQCHKYCLPSSLKLLTSLQFSHSSSLSLATHKRVRTRTSVVAMA
Sbjct: 168 MGMATRVPFVSATLSPNQCHKYCLPSSLKLLTSLQFSHSSSLSLATHKRVRTRTSVVAMA 347
Query: 61 DSSRSTVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTIGASDDVYVGDIRDT 120
DSSRSTVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTIGASDDVYVGDIRDT
Sbjct: 348 DSSRSTVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTIGASDDVYVGDIRDT 527
Query: 121 GSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKN 175
GSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKN
Sbjct: 528 GSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWIGQKN 692
>TC14303 similar to UP|Y230_ARATH (O80934) Protein At2g37660, chloroplast
precursor, partial (42%)
Length = 721
Score = 270 bits (690), Expect = 2e-73
Identities = 136/137 (99%), Positives = 137/137 (99%)
Frame = +2
Query: 177 IDAAKAAGVKQIVLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGG 236
IDAAKAAGV+QIVLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGG
Sbjct: 2 IDAAKAAGVRQIVLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGG 181
Query: 237 LQDKEGGVRELIIGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDLASKPEGAG 296
LQDKEGGVRELIIGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDLASKPEGAG
Sbjct: 182 LQDKEGGVRELIIGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDLASKPEGAG 361
Query: 297 TPTRDFKALFSQITTRF 313
TPTRDFKALFSQITTRF
Sbjct: 362 TPTRDFKALFSQITTRF 412
>TC9101 similar to GB|AAL08281.1|15912295|AY056425 At2g34460/T31E10.20
{Arabidopsis thaliana;}, partial (49%)
Length = 637
Score = 58.2 bits (139), Expect = 2e-09
Identities = 43/126 (34%), Positives = 65/126 (51%), Gaps = 2/126 (1%)
Frame = +1
Query: 189 VLVGSMGGTDLNNPLNSLGN--GNILVWKRKAEQYLADSGIPYTIIRAGGLQDKEGGVRE 246
+LV L NP N G LV K +AE ++ SGI YTIIR GGL++ +
Sbjct: 31 ILVNGAAMGQLLNPAYIFLNVFGLTLVAKLQAENHIRKSGINYTIIRPGGLRN-DPPTGN 207
Query: 247 LIIGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDLASKPEGAGTPTRDFKALF 306
+++ +D + +I+R VAEV ++AL EA +K ++ S+P+ P R + LF
Sbjct: 208 VVMEPEDTL---SQGSISRDHVAEVAVEALACPEASYKVVEIVSRPD---APKRTYHDLF 369
Query: 307 SQITTR 312
I R
Sbjct: 370 GSIRQR 387
>TC11437 homologue to GB|AAP37706.1|30725368|BT008347 At4g31530 {Arabidopsis
thaliana;}, partial (27%)
Length = 552
Score = 55.8 bits (133), Expect = 9e-09
Identities = 35/111 (31%), Positives = 61/111 (54%), Gaps = 15/111 (13%)
Frame = -2
Query: 211 ILVWKRKAEQYLADSGIPYTIIRAGGLQD--------------KEGGVRELIIGKDDEIL 256
+L +K+ E +L +SGIP+TIIR G L D G R ++IG+ D+++
Sbjct: 515 VLKYKKMGEDFLQNSGIPFTIIRPGRLTDGPYTSYDLNTLLKATAGQRRAVLIGQGDKLV 336
Query: 257 KTETRTIARPDVAEVCIQALNFEEAQFKAFDLAS-KPEGAGTPTRDFKALF 306
+R + VAE CIQAL+ + + + ++++S + EG G + ++ LF
Sbjct: 335 GEASRIV----VAEACIQALDLQVTENQVYEVSSVEGEGPGNEAQKWEELF 195
>TC11937 weakly similar to UP|AAS49102 (AAS49102) At1g72640, partial (60%)
Length = 774
Score = 52.0 bits (123), Expect = 1e-07
Identities = 30/93 (32%), Positives = 46/93 (49%)
Frame = +3
Query: 184 GVKQIVLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQYLADSGIPYTIIRAGGLQDKEGG 243
GV+ ++ + + + + S+ N N + E L SGIPYTIIR G LQD GG
Sbjct: 228 GVQHVIFLSQLSVYSGKSGIQSMMNSNARKLAEQDESALVTSGIPYTIIRTGALQDTPGG 407
Query: 244 VRELIIGKDDEILKTETRTIARPDVAEVCIQAL 276
+ K + +I++ D A VC++AL
Sbjct: 408 KQGFTFDKG----SAASGSISKEDAAFVCVEAL 494
>TC16256 similar to UP|Q7Y0H8 (Q7Y0H8) Cinnamoyl CoA reductase, partial
(32%)
Length = 475
Score = 38.9 bits (89), Expect = 0.001
Identities = 37/123 (30%), Positives = 56/123 (45%), Gaps = 16/123 (13%)
Frame = +3
Query: 34 LQFSHSSSLSLATH-----KRVRTRTSVVAMADSSRST---VLVTGAGGRTGKIVYKKLQ 85
LQ S+++S +H K+++ + A AD+S + V VTGAGG + K L
Sbjct: 21 LQILFSTTISFFSHLYNQRKKLKESKKMPAPADTSSVSGDIVCVTGAGGFIASWLVKLLL 200
Query: 86 ERSSQYIARGLVRTEES------KQTIGASD--DVYVGDIRDTGSIAPAIQGIDALIILT 137
ER YI RG VR + K+ GA + ++ D+ D S+ G D +
Sbjct: 201 ERG--YIVRGTVRNPDDPKNGHLKELEGAKERLTLHKADLFDLDSLKAVFHGCDGVFHTA 374
Query: 138 SAV 140
S V
Sbjct: 375 SPV 383
>TC7966 similar to UP|Q84V25 (Q84V25) Quinone oxidoreductase, partial (98%)
Length = 1908
Score = 38.1 bits (87), Expect = 0.002
Identities = 41/156 (26%), Positives = 66/156 (42%), Gaps = 10/156 (6%)
Frame = -1
Query: 50 VRTRTSVVA--MADSSRSTVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEES----- 102
+R+ T++ M ++ T+ VTGAGG + K L ++ Y RG VR +
Sbjct: 645 IRSHTTITNDNMPAAAGETICVTGAGGFIASWMVKLLLQKG--YTVRGTVRNPDDPKNGH 472
Query: 103 -KQTIGASDDVYV--GDIRDTGSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYF 159
++ GASD + + D+ + S+ A+ G + S V + PE
Sbjct: 471 LRELEGASDRLTLIKVDLLELNSVRAAVHGSHGVFHTASPVT-----------DNPEEMV 325
Query: 160 EDGAYPEQVDWIGQKNQIDAAKAAGVKQIVLVGSMG 195
E G KN I AA A V+++V S+G
Sbjct: 324 EPAVN-------GAKNVIIAAAEAKVRRVVFTSSIG 238
>TC17328 weakly similar to UP|Q9FGH3 (Q9FGH3) Dihydroflavonol
4-reductase-like (Cinnamoyl-CoA reductase-like protein),
partial (21%)
Length = 479
Score = 28.5 bits (62), Expect = 1.5
Identities = 22/83 (26%), Positives = 38/83 (45%)
Frame = +3
Query: 112 VYVGDIRDTGSIAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYFEDGAYPEQVDWI 171
++ DI D+ +++ AI + + S L + PE +D P
Sbjct: 84 IFPADILDSAAVSRAIHNCSGVFHVASPCTL----------DDPEDPHKDLIQPAVQ--- 224
Query: 172 GQKNQIDAAKAAGVKQIVLVGSM 194
G N ++AAK AGV+++VL S+
Sbjct: 225 GTLNVLEAAKRAGVRRVVLTSSI 293
>TC13337
Length = 531
Score = 27.7 bits (60), Expect = 2.6
Identities = 15/44 (34%), Positives = 28/44 (63%)
Frame = +3
Query: 245 RELIIGKDDEILKTETRTIARPDVAEVCIQALNFEEAQFKAFDL 288
R LI + + I ++ I+ DVA++C++AL+ A+ K+FD+
Sbjct: 3 RALIFDQGNRI----SQGISCADVADICVKALHDSTARNKSFDV 122
>TC7816 homologue to UP|Q9ATM4 (Q9ATM4) Plasma membrane integral protein
ZmPIP2-7, complete
Length = 1523
Score = 26.9 bits (58), Expect = 4.5
Identities = 15/52 (28%), Positives = 25/52 (47%), Gaps = 2/52 (3%)
Frame = +1
Query: 123 IAPAIQGIDALIILTSAVPLMKPGFNPTKGERPEFYF-EDGAYPEQ-VDWIG 172
+AP G ++ + +P+ G NP + P F ED + +Q V W+G
Sbjct: 784 LAPLPIGFAVFMVHLATIPVTGTGINPARSFGPAVIFNEDKVWDDQWVYWVG 939
>TC8218 homologue to UP|Q93VR3 (Q93VR3) AT5g28840/F7P1_20, complete
Length = 1590
Score = 26.6 bits (57), Expect = 5.8
Identities = 16/77 (20%), Positives = 35/77 (44%)
Frame = +3
Query: 62 SSRSTVLVTGAGGRTGKIVYKKLQERSSQYIARGLVRTEESKQTIGASDDVYVGDIRDTG 121
S + + +TGAGG + ++L+ IA + E + + D+ ++ D+R
Sbjct: 138 SEKLKISITGAGGFIASHIARRLKTEGHYIIASDWKKNEHMSEDM-FCDEFHLVDLRVMD 314
Query: 122 SIAPAIQGIDALIILTS 138
+ +G+D + L +
Sbjct: 315 NCLKVTKGVDHVFNLAA 365
>TC19468 similar to UP|Q7VB79 (Q7VB79) Predicted protein family PM-3,
partial (9%)
Length = 530
Score = 26.6 bits (57), Expect = 5.8
Identities = 32/132 (24%), Positives = 57/132 (42%), Gaps = 9/132 (6%)
Frame = +1
Query: 35 QFSHSSSLSLATHK----RVRTRTSVVAMADSSRSTVLVTGAGGRTGK---IVYKKLQER 87
+FS S+L T K R+ +V S ST+ +T A T + I K
Sbjct: 49 KFSKFSTLGFPTFKTEFKRLFQIEAVSRRFPMSASTLSITAANPGTRRRPVIAADKKTAS 228
Query: 88 SSQYIARGLVRTEES--KQTIGASDDVYVGDIRDTGSIAPAIQGIDALIILTSAVPLMKP 145
+ + +A + + + +T A D+ IR + PA + + ++AVP+ +
Sbjct: 229 NLELLANDVAVSPAAVDAKTPSAGRDLSHHSIRGEAYLEPAAKTV------STAVPVRRA 390
Query: 146 GFNPTKGERPEF 157
+PTK E+P +
Sbjct: 391 RKSPTKAEKPRW 426
>TC17786 similar to PIR|T46177|T46177 villin 3 homolog T8H10.10 -
Arabidopsis thaliana (fragment) {Arabidopsis thaliana;},
partial (42%)
Length = 1228
Score = 26.6 bits (57), Expect = 5.8
Identities = 18/59 (30%), Positives = 25/59 (41%), Gaps = 13/59 (22%)
Frame = +3
Query: 151 KGERPEFYFED------------GAYPEQVD-WIGQKNQIDAAKAAGVKQIVLVGSMGG 196
K E +FY D G+Y + WIG+ D A A +K + L S+GG
Sbjct: 408 KSEHGKFYMGDSYIILQTTQGKGGSYLFDIHFWIGKDTSQDEAGTAAIKTVELDASLGG 584
>TC8417 similar to UP|GAE1_PEA (Q43070) UDP-glucose 4-epimerase
(Galactowaldenase) (UDP-galactose 4-epimerase) ,
partial (65%)
Length = 789
Score = 26.6 bits (57), Expect = 5.8
Identities = 13/42 (30%), Positives = 22/42 (51%)
Frame = +3
Query: 36 FSHSSSLSLATHKRVRTRTSVVAMADSSRSTVLVTGAGGRTG 77
F S SL + TH+ + ++ + + S + +LVTG G G
Sbjct: 18 FQFSRSL*ILTHQILSPNSNKMVSSSSPQQKILVTGGAGFIG 143
>TC9535
Length = 1083
Score = 26.2 bits (56), Expect = 7.6
Identities = 17/57 (29%), Positives = 27/57 (46%), Gaps = 6/57 (10%)
Frame = +3
Query: 189 VLVGSMGGTDLNNPLNSLGNGNILVWKRKAEQY------LADSGIPYTIIRAGGLQD 239
V G +GG +L+ + +G++ V ++E SGIP RAGG+ D
Sbjct: 384 VFTGMLGGEELSQ---AYASGDVFVMPSESETLGLVVLEAMSSGIPVVGARAGGIPD 545
>TC19596
Length = 499
Score = 26.2 bits (56), Expect = 7.6
Identities = 16/54 (29%), Positives = 25/54 (45%)
Frame = +1
Query: 161 DGAYPEQVDWIGQKNQIDAAKAAGVKQIVLVGSMGGTDLNNPLNSLGNGNILVW 214
DGA+ D +G + A A + GS+GG L + +L +G L+W
Sbjct: 16 DGAFKHDDDRMGMGGIVRDAHGAWISGFY-AGSLGGDALRAEIAALKHGLTLLW 174
>TC8724 homologue to UP|Q9AQU5 (Q9AQU5) Plasma membrane integral protein
ZmPIP1-4 (Plasma membrane integral protein ZmPIP1-3),
partial (93%)
Length = 1560
Score = 25.8 bits (55), Expect = 9.9
Identities = 20/70 (28%), Positives = 32/70 (45%), Gaps = 2/70 (2%)
Frame = +1
Query: 123 IAPAIQGIDALIILTSAVPLMKPGFNPTKG-ERPEFYFEDGAYPEQ-VDWIGQKNQIDAA 180
+AP G ++ + +P+ G NP + Y D A+ + + W+G I AA
Sbjct: 718 LAPLPIGFAVFLVHLATIPITGTGINPARSLGAAIIYNRDHAWDDHWIFWVGP--FIGAA 891
Query: 181 KAAGVKQIVL 190
AA QIV+
Sbjct: 892 LAAVYHQIVI 921
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.134 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,281,149
Number of Sequences: 28460
Number of extensions: 50215
Number of successful extensions: 325
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 319
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 321
length of query: 313
length of database: 4,897,600
effective HSP length: 90
effective length of query: 223
effective length of database: 2,336,200
effective search space: 520972600
effective search space used: 520972600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0147b.4


