

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
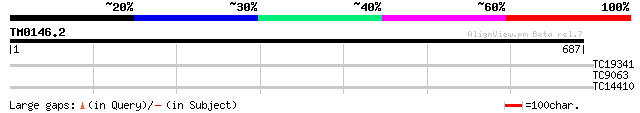
Query= TM0146.2
(687 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC19341 similar to UP|Q9CAD1 (Q9CAD1) GMP synthase, partial (28%) 30 1.3
TC9063 similar to UP|Q8LFK9 (Q8LFK9) 3-dehydroquinate synthase-l... 27 8.4
TC14410 homologue to UP|Q7Y0W1 (Q7Y0W1) GL2-type homeodomain pro... 27 8.4
>TC19341 similar to UP|Q9CAD1 (Q9CAD1) GMP synthase, partial (28%)
Length = 484
Score = 30.0 bits (66), Expect = 1.3
Identities = 28/86 (32%), Positives = 41/86 (47%), Gaps = 2/86 (2%)
Frame = +1
Query: 391 NSYPNASDSALPKEHHITLWRTLLS-PRVLTVGFASSDYTLCGYNPQL-VSRQFGLSQVL 448
+S +A+ SAL + +WR+LLS PR LC +L +S+ FG + V+
Sbjct: 232 SSSASAAMSALDTRRSMVVWRSLLSTPR------------LCSPPTRLAISKLFG*AMVM 375
Query: 449 PNTLFDKSLVLYPGTIKRVSVFDKTV 474
LF +L PG K S KT+
Sbjct: 376 KPLLFLLVSMLLPGVSKAPSPPSKTL 453
>TC9063 similar to UP|Q8LFK9 (Q8LFK9) 3-dehydroquinate synthase-like
protein, partial (14%)
Length = 586
Score = 27.3 bits (59), Expect = 8.4
Identities = 11/30 (36%), Positives = 16/30 (52%)
Frame = -3
Query: 111 PNPSANDKAYLKWLDRVEGDKKQHWKDVGI 140
P+ N K L W D+ G K+ W D+G+
Sbjct: 293 PHLGFNRKWVLGWEDKAWGGAKERWVDLGV 204
>TC14410 homologue to UP|Q7Y0W1 (Q7Y0W1) GL2-type homeodomain protein,
partial (4%)
Length = 780
Score = 27.3 bits (59), Expect = 8.4
Identities = 19/53 (35%), Positives = 24/53 (44%), Gaps = 3/53 (5%)
Frame = -2
Query: 471 DKTVQFYNKKLLNLSPFTYAPSY---YATNAFKAWWSEYWAQISKPLVDCLQC 520
+KT N L N SPF S+ AT K+W ISKPL ++C
Sbjct: 710 EKTEHMINVALSNSSPFFLG*SHGSKRATPYMKSWAKWKQTPISKPLKGQIRC 552
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.136 0.427
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,903,144
Number of Sequences: 28460
Number of extensions: 213075
Number of successful extensions: 1092
Number of sequences better than 10.0: 6
Number of HSP's better than 10.0 without gapping: 1088
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1092
length of query: 687
length of database: 4,897,600
effective HSP length: 96
effective length of query: 591
effective length of database: 2,165,440
effective search space: 1279775040
effective search space used: 1279775040
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0146.2


