

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0141.9
(378 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
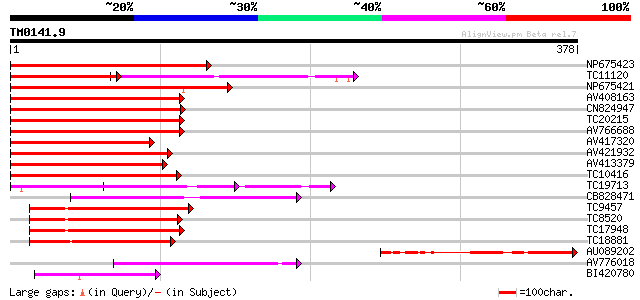
Score E
Sequences producing significant alignments: (bits) Value
NP675423 transcription factor MYB101 [Lotus japonicus] 230 3e-61
TC11120 UP|Q84PP4 (Q84PP4) Transcription factor MYB102, complete 140 2e-59
NP675421 transcription factor MYB103 [Lotus japonicus] 204 3e-53
AV408163 180 4e-46
CN824947 178 2e-45
TC20215 similar to AAS10104 (AAS10104) MYB transcription factor,... 176 6e-45
AV766688 164 2e-41
AV417320 162 7e-41
AV421932 162 9e-41
AV413379 161 2e-40
TC10416 weakly similar to UP|O04108 (O04108) MYB factor, partial... 158 1e-39
TC19713 homologue to UP|P93474 (P93474) Myb26, partial (55%) 141 2e-34
CB828471 132 7e-32
TC9457 homologue to UP|Q9FZ15 (Q9FZ15) Tuber-specific and sucros... 114 3e-26
TC8520 homologue to UP|Q9FZ15 (Q9FZ15) Tuber-specific and sucros... 112 1e-25
TC17948 similar to UP|Q9FZ13 (Q9FZ13) Tuber-specific and sucrose... 111 2e-25
TC18881 similar to UP|Q9FZ15 (Q9FZ15) Tuber-specific and sucrose... 106 7e-24
AU089202 104 3e-23
AV776018 86 1e-17
BI420780 72 1e-13
>NP675423 transcription factor MYB101 [Lotus japonicus]
Length = 1047
Score = 230 bits (586), Expect = 3e-61
Identities = 100/134 (74%), Positives = 120/134 (88%)
Frame = +1
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGR+PCC+++GLKKGPWTPEED L+ YI++HGHGSWRALP AGL RCGKSCRLRW+NY
Sbjct: 1 MGRTPCCDEIGLKKGPWTPEEDAILVDYIQKHGHGSWRALPKLAGLNRCGKSCRLRWTNY 180
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMG 120
LRPDIKRGKF+ +EEQ II LHA+LGN+WSAIA HL RTDNEIKN+WNTHLKK+L++MG
Sbjct: 181 LRPDIKRGKFTEEEEQLIINLHAVLGNKWSAIAGHLPGRTDNEIKNFWNTHLKKKLLQMG 360
Query: 121 IDPVTHKPKNDALH 134
+DPVTH+P+ D L+
Sbjct: 361 LDPVTHRPRLDHLN 402
>TC11120 UP|Q84PP4 (Q84PP4) Transcription factor MYB102, complete
Length = 1281
Score = 140 bits (352), Expect(2) = 2e-59
Identities = 60/74 (81%), Positives = 68/74 (91%)
Frame = +1
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGRSPCC++ GLKKGPWTPEEDQKL+ +I++HGHGSWRALP AGL RCGKSCRLRW+NY
Sbjct: 67 MGRSPCCDENGLKKGPWTPEEDQKLVEHIQKHGHGSWRALPKLAGLNRCGKSCRLRWTNY 246
Query: 61 LRPDIKRGKFSLQE 74
LRPDIKRGKFS +E
Sbjct: 247 LRPDIKRGKFSPEE 288
Score = 105 bits (263), Expect(2) = 2e-59
Identities = 63/172 (36%), Positives = 101/172 (58%), Gaps = 7/172 (4%)
Frame = +2
Query: 68 GKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMGIDPVTHK 127
G F ++EQTI+ LH++LGN+WS IATHL RTDNEIKN+WNTHLKK+LI+MG DP+TH+
Sbjct: 269 GNFLQKKEQTILHLHSILGNKWSTIATHLPGRTDNEIKNFWNTHLKKKLIQMGFDPMTHQ 448
Query: 128 PKNDALHSSIDNGHSKVAANLSHMAQWESARLEAEA-RLVRESKLRSHLGTSSSILPSPS 186
P+ D + + + AN++ + + +A +L + L L +S++ L +
Sbjct: 449 PRTDLVSTL---PYLLALANMTQLMDHNHQSFDEQAVQLAKLQYLNYLLQSSNNPLCMTN 619
Query: 187 SSSLNKHDTVKAWNNGSSVVGDLESPKSTL--SFSENNNN----NHNDNNVA 232
S N ++A+ S++ + + K L FS+ + +H+ NN+A
Sbjct: 620 SYDQNALTNMEAF----SLLNSISNVKENLGVDFSQMDTQTSLFSHDGNNMA 763
>NP675421 transcription factor MYB103 [Lotus japonicus]
Length = 924
Score = 204 bits (518), Expect = 3e-53
Identities = 95/150 (63%), Positives = 119/150 (79%), Gaps = 2/150 (1%)
Frame = +1
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
M R+P ++ G+KKGPW+ EED+ L+ YI++HGHGSWRALP AGL RCGKSCRLRW+NY
Sbjct: 1 MMRTPSIDESGMKKGPWSQEEDKVLVDYIQKHGHGSWRALPQLAGLNRCGKSCRLRWTNY 180
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKK--RLIK 118
LRPDIKRGKFS +EEQ II LHA LGN+W+ IA+HL RTDNEIKN WNTHLKK +L++
Sbjct: 181 LRPDIKRGKFSDEEEQLIINLHASLGNKWATIASHLPGRTDNEIKNLWNTHLKKKLKLLQ 360
Query: 119 MGIDPVTHKPKNDALHSSIDNGHSKVAANL 148
MG+DPVTH P++D ++ + AAN+
Sbjct: 361 MGLDPVTHCPRSDHMNILSNLQQILAAANI 450
>AV408163
Length = 425
Score = 180 bits (456), Expect = 4e-46
Identities = 77/116 (66%), Positives = 94/116 (80%)
Frame = +2
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGRSPCCEK + KG WT EED +L++YI HG G WR+LP AGL RCGKSCRLRW NY
Sbjct: 77 MGRSPCCEKAHINKGAWTKEEDDRLISYIRAHGEGCWRSLPKAAGLLRCGKSCRLRWINY 256
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRL 116
LRPD+KRG F+ +E++ II+LH+LLGN+WS IA L RTDNEIKNYWNTH++++L
Sbjct: 257 LRPDLKRGNFTEEEDELIIKLHSLLGNKWSLIAGRLPGRTDNEIKNYWNTHIRRKL 424
>CN824947
Length = 738
Score = 178 bits (451), Expect = 2e-45
Identities = 78/117 (66%), Positives = 93/117 (78%)
Frame = +1
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGR+PCCEKVGLK+G WT EED+ L YI+ G GSWR+LP AGL RCGKSCRLRW NY
Sbjct: 151 MGRAPCCEKVGLKRGRWTAEEDELLTKYIQASGEGSWRSLPKNAGLLRCGKSCRLRWINY 330
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLI 117
LR D+KRG S +EE I++LHA GNRWS IA+H+ RTDNEIKNYWN+HL ++L+
Sbjct: 331 LRGDLKRGNISDEEESLIVKLHASFGNRWSLIASHMPGRTDNEIKNYWNSHLSRKLV 501
>TC20215 similar to AAS10104 (AAS10104) MYB transcription factor, partial
(35%)
Length = 464
Score = 176 bits (446), Expect = 6e-45
Identities = 75/116 (64%), Positives = 96/116 (82%)
Frame = +2
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGR+PCCEKVGLK+G W+ EED+ L +I+++G GSW++LP AGL RCGKSCRLRW NY
Sbjct: 29 MGRAPCCEKVGLKRGRWSEEEDKILTDFIKQNGEGSWKSLPKNAGLLRCGKSCRLRWINY 208
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRL 116
LR D+KRG + +EE+ I++LHA LGNRWS IA HL RTDNEIKNYWN+HL++++
Sbjct: 209 LREDVKRGNITSEEEEIIVKLHAALGNRWSVIAGHLPGRTDNEIKNYWNSHLRRKI 376
>AV766688
Length = 608
Score = 164 bits (416), Expect = 2e-41
Identities = 73/116 (62%), Positives = 91/116 (77%)
Frame = +2
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
M R+P C+K GL+KG WT EED+KL+AY+ +G +WR LP AGL RCGKSCRLRW NY
Sbjct: 77 MARTPSCDKSGLRKGTWTAEEDRKLIAYVTRYGCWNWRQLPKFAGLARCGKSCRLRWLNY 256
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRL 116
LRPDIKRG F+ +EE+ II++H LGNRWS IA L RTDNE+KN+W+T LK+R+
Sbjct: 257 LRPDIKRGHFTQEEEEIIIRMHKNLGNRWSIIAAELPGRTDNEVKNHWHTSLKRRV 424
>AV417320
Length = 369
Score = 162 bits (411), Expect = 7e-41
Identities = 71/96 (73%), Positives = 83/96 (85%)
Frame = +3
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
M R+PCCEK+GLKKGPWT EEDQ L++YI++HGHG+WRALP AGL RCGKSCRLRW NY
Sbjct: 78 MVRAPCCEKIGLKKGPWTSEEDQILISYIQKHGHGNWRALPKHAGLLRCGKSCRLRWINY 257
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHL 96
LRPDIKRG F+ +EE+ II++H LLGNRWSAIA L
Sbjct: 258 LRPDIKRGNFTAEEEELIIKMHELLGNRWSAIAAKL 365
>AV421932
Length = 402
Score = 162 bits (410), Expect = 9e-41
Identities = 69/108 (63%), Positives = 86/108 (78%)
Frame = +3
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MG CC + +K+G W+PEED+KL+ YI HG+G W +P KAGLQRCGKSCRLRW NY
Sbjct: 78 MGHHSCCNQQKVKRGLWSPEEDEKLIRYITTHGYGCWSEVPEKAGLQRCGKSCRLRWINY 257
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYW 108
LRPDI+RG+F+ +EE+ II LH ++GNRW+ IA+HL RTDNEIKNYW
Sbjct: 258 LRPDIRRGRFTPEEEKLIISLHGVVGNRWAHIASHLPGRTDNEIKNYW 401
>AV413379
Length = 416
Score = 161 bits (407), Expect = 2e-40
Identities = 72/105 (68%), Positives = 83/105 (78%)
Frame = +3
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
MGRSPCCEK KG WT EED +L+AYI HG G WR+LP AGL RCGKSCRLRW NY
Sbjct: 102 MGRSPCCEKAHTNKGAWTKEEDDRLIAYIRAHGEGCWRSLPKSAGLLRCGKSCRLRWINY 281
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIK 105
LRPD+KRG F+ QE++ II+LH+LLGN+WS IA L+ RTDNEIK
Sbjct: 282 LRPDLKRGNFTPQEDELIIKLHSLLGNKWSLIAGRLAGRTDNEIK 416
>TC10416 weakly similar to UP|O04108 (O04108) MYB factor, partial (41%)
Length = 1046
Score = 158 bits (400), Expect = 1e-39
Identities = 69/114 (60%), Positives = 89/114 (77%)
Frame = +2
Query: 1 MGRSPCCEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNY 60
M R+P C+K G++KG WT EED+KL+AY+ +G +WR LP AGL RCGKSCRLRW NY
Sbjct: 92 MARTPSCDKSGMRKGTWTAEEDKKLIAYVTRYGCWNWRQLPKFAGLARCGKSCRLRWLNY 271
Query: 61 LRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKK 114
LRP+IKRG F+ +EE+ II++H LGNRWS IA L RTD+E+KN+W+T LK+
Sbjct: 272 LRPNIKRGNFTQEEEEIIIRMHKKLGNRWSTIAAELPGRTDSEVKNHWHTTLKR 433
>TC19713 homologue to UP|P93474 (P93474) Myb26, partial (55%)
Length = 532
Score = 141 bits (355), Expect = 2e-34
Identities = 70/155 (45%), Positives = 93/155 (59%), Gaps = 2/155 (1%)
Frame = +3
Query: 1 MGRSPC--CEKVGLKKGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWS 58
M + PC ++KGPWT EED L+ YI HG G W L AGL+R GKSCRLRW
Sbjct: 66 MDKKPCNSSHDPEVRKGPWTIEEDLILINYIANHGEGVWNTLAKSAGLKRTGKSCRLRWL 245
Query: 59 NYLRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIK 118
NYLRPD++RG + +E+ I++LHA GNRWS IA HL RTDNEIKN+W T ++K + +
Sbjct: 246 NYLRPDVRRGNITPEEQLLIMELHAKWGNRWSKIAKHLPGRTDNEIKNFWRTRIQKHIKQ 425
Query: 119 MGIDPVTHKPKNDALHSSIDNGHSKVAANLSHMAQ 153
+ S I++ H + +S MA+
Sbjct: 426 ------AESLQQQQSSSEINDHHQTSTSQMSTMAE 512
Score = 40.0 bits (92), Expect = 6e-04
Identities = 35/157 (22%), Positives = 69/157 (43%), Gaps = 2/157 (1%)
Frame = +3
Query: 63 PDIKRGKFSLQEEQTIIQLHALLGNR-WSAIATHLS-KRTDNEIKNYWNTHLKKRLIKMG 120
P++++G ++++E+ +I A G W+ +A KRT + W +L+ + +
Sbjct: 99 PEVRKGPWTIEEDLILINYIANHGEGVWNTLAKSAGLKRTGKSCRLRWLNYLRPDVRRGN 278
Query: 121 IDPVTHKPKNDALHSSIDNGHSKVAANLSHMAQWESARLEAEARLVRESKLRSHLGTSSS 180
I P + LH+ N SK+A +L R + E + ++++ H+ + S
Sbjct: 279 ITPEEQLLIME-LHAKWGNRWSKIAKHLP-------GRTDNEIKNFWRTRIQKHIKQAES 434
Query: 181 ILPSPSSSSLNKHDTVKAWNNGSSVVGDLESPKSTLS 217
+ SSS +N H +S + + P T S
Sbjct: 435 LQQQQSSSEINDHH-----QTSTSQMSTMAEPMDTYS 530
>CB828471
Length = 525
Score = 132 bits (333), Expect = 7e-32
Identities = 74/154 (48%), Positives = 90/154 (58%)
Frame = +1
Query: 41 PAKAGLQRCGKSCRLRWSNYLRPDIKRGKFSLQEEQTIIQLHALLGNRWSAIATHLSKRT 100
P +AGL RCGKSCRLRW+NYLRPDIKRG FS +EE II LH LLGNRWSAIA L RT
Sbjct: 1 PKQAGLLRCGKSCRLRWANYLRPDIKRGNFSREEEDEIINLHELLGNRWSAIAARLPGRT 180
Query: 101 DNEIKNYWNTHLKKRLIKMGIDPVTHKPKNDALHSSIDNGHSKVAANLSHMAQWESARLE 160
DNEIKN W+THLKKRL +P+ +++D SK + Q +
Sbjct: 181 DNEIKNVWHTHLKKRL----------QPQTKQKQTNLDLDASKSNKDAKKEHQEDEGPRS 330
Query: 161 AEARLVRESKLRSHLGTSSSILPSPSSSSLNKHD 194
+ S S L SSSI PS + ++ +D
Sbjct: 331 PQQCSSSNSDNMSSLTNSSSIAPSTNHDDMSIND 432
>TC9457 homologue to UP|Q9FZ15 (Q9FZ15) Tuber-specific and
sucrose-responsive element binding factor, partial (43%)
Length = 860
Score = 114 bits (284), Expect = 3e-26
Identities = 53/109 (48%), Positives = 74/109 (67%)
Frame = +3
Query: 14 KGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNYLRPDIKRGKFSLQ 73
KGPW+PEED+ L +E+HG +W +L +K+ R GKSCRLRW N L P ++ F+ +
Sbjct: 78 KGPWSPEEDEALQRLVEKHGPRNW-SLISKSIPGRSGKSCRLRWCNQLSPQVEHRAFTAE 254
Query: 74 EEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMGID 122
E+ TII+ HA GN+W+ IA LS RTDN IKN+WN+ LK++ +D
Sbjct: 255 EDDTIIRAHARFGNKWATIARLLSGRTDNAIKNHWNSTLKRKCASFMMD 401
>TC8520 homologue to UP|Q9FZ15 (Q9FZ15) Tuber-specific and
sucrose-responsive element binding factor, partial (36%)
Length = 1778
Score = 112 bits (279), Expect = 1e-25
Identities = 52/102 (50%), Positives = 71/102 (68%)
Frame = +1
Query: 14 KGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNYLRPDIKRGKFSLQ 73
KGPW+PEED+ L +E HG +W +L +K+ R GKSCRLRW N L P ++ F+ +
Sbjct: 184 KGPWSPEEDEALQKLVERHGPRNW-SLISKSIPGRSGKSCRLRWCNQLSPQVEHRAFTPE 360
Query: 74 EEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKR 115
E+ TII+ HA GN+W+ IA LS RTDN IKN+WN+ LK++
Sbjct: 361 EDDTIIRAHARFGNKWATIARLLSGRTDNAIKNHWNSTLKRK 486
>TC17948 similar to UP|Q9FZ13 (Q9FZ13) Tuber-specific and sucrose-responsive
element binding factor, partial (50%)
Length = 487
Score = 111 bits (278), Expect = 2e-25
Identities = 53/103 (51%), Positives = 71/103 (68%)
Frame = +2
Query: 14 KGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNYLRPDIKRGKFSLQ 73
KGPW+ EED+ L +E HG +W +L ++ R GKSCRLRW N L P ++ FS+Q
Sbjct: 170 KGPWSAEEDRILTRLVERHGARNW-SLISRYIKGRSGKSCRLRWCNQLSPTVEHRPFSVQ 346
Query: 74 EEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRL 116
E++TII HA GNRW+AIA L RTDN +KN+WN+ LK+R+
Sbjct: 347 EDETIIAAHARFGNRWAAIARLLPGRTDNAVKNHWNSTLKRRV 475
>TC18881 similar to UP|Q9FZ15 (Q9FZ15) Tuber-specific and sucrose-responsive
element binding factor, partial (28%)
Length = 346
Score = 106 bits (264), Expect = 7e-24
Identities = 47/97 (48%), Positives = 68/97 (69%)
Frame = +1
Query: 14 KGPWTPEEDQKLLAYIEEHGHGSWRALPAKAGLQRCGKSCRLRWSNYLRPDIKRGKFSLQ 73
KGPW+PEED+ L ++ HG +W A+ +K+ R GKSCRLRW N L P+++ F+ +
Sbjct: 55 KGPWSPEEDEALTRLVQVHGPKNWTAI-SKSIPGRSGKSCRLRWCNQLSPEVEHRPFTPE 231
Query: 74 EEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNT 110
E+ TII+ HA GN+W+ IA L+ RTDN +KN+WN+
Sbjct: 232 EDSTIIRAHARCGNKWATIARILNGRTDNAVKNHWNS 342
>AU089202
Length = 401
Score = 104 bits (259), Expect = 3e-23
Identities = 70/132 (53%), Positives = 81/132 (61%), Gaps = 1/132 (0%)
Frame = +3
Query: 248 VKEEGEQEWKGYKDGMENSMPFTSTTFHELTMTMEATWPPSDESLRTITTAATTAARAHV 307
VKEEGEQ WKGY E+SM FTS FHE ME V
Sbjct: 9 VKEEGEQ-WKGYH---ESSMTFTSG-FHEF---MEG-----------------------V 95
Query: 308 LGDPVVQEGLTDLLLNSNSDEDRSLSPEGGGESNSGDGGDGCGSDFYE-DNKNYWNNILN 366
+GD VV+EG T+LLL +NSD+ EGGGESN+G G G GSDFYE DN NYW++ILN
Sbjct: 96 VGDDVVEEGFTNLLLKTNSDDS-----EGGGESNNG--GRGSGSDFYEADNNNYWSSILN 254
Query: 367 LVNSSPTNSPMF 378
LVNSSP++SPMF
Sbjct: 255 LVNSSPSDSPMF 290
>AV776018
Length = 476
Score = 85.5 bits (210), Expect = 1e-17
Identities = 51/126 (40%), Positives = 72/126 (56%), Gaps = 1/126 (0%)
Frame = +1
Query: 70 FSLQEEQTIIQLHALLGNRWSAIATHLSKRTDNEIKNYWNTHLKKRLIKMGIDPVTHKPK 129
F +E+ II+LHA+LGNRWS AT RTDNEIKN WN+ LKK+L + GIDPVTHKP
Sbjct: 13 FHKEEDNLIIELHAVLGNRWSQDATQWPGRTDNEIKNLWNSCLKKKLRQRGIDPVTHKPL 192
Query: 130 NDALHSSID-NGHSKVAANLSHMAQWESARLEAEARLVRESKLRSHLGTSSSILPSPSSS 188
++ + D N + +N ++ + ES + +A + S L T+ + SSS
Sbjct: 193 SEVENGKEDKNKSQEKVSNELNLLKSESFKSDAASYEQNSSSLAPKAYTTE--MEGSSSS 366
Query: 189 SLNKHD 194
+ D
Sbjct: 367 DCSSKD 384
>BI420780
Length = 475
Score = 72.4 bits (176), Expect = 1e-13
Identities = 34/86 (39%), Positives = 50/86 (57%), Gaps = 2/86 (2%)
Frame = +1
Query: 17 WTPEEDQKLLAYIEEHGHGSWRALPAKAG--LQRCGKSCRLRWSNYLRPDIKRGKFSLQE 74
W+ EED L AY++++G W + + L R KSC RW NYL+P IK+G + +E
Sbjct: 211 WSSEEDALLHAYVQQYGPREWNLVSQRMNTPLNRDTKSCLERWKNYLKPGIKKGSLTKEE 390
Query: 75 EQTIIQLHALLGNRWSAIATHLSKRT 100
++ +I L A GN+W IA + RT
Sbjct: 391 QRLVILLQANYGNKWKKIAAEVPGRT 468
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.310 0.128 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,458,777
Number of Sequences: 28460
Number of extensions: 112315
Number of successful extensions: 868
Number of sequences better than 10.0: 80
Number of HSP's better than 10.0 without gapping: 742
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 812
length of query: 378
length of database: 4,897,600
effective HSP length: 92
effective length of query: 286
effective length of database: 2,279,280
effective search space: 651874080
effective search space used: 651874080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0141.9


