

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0137.11
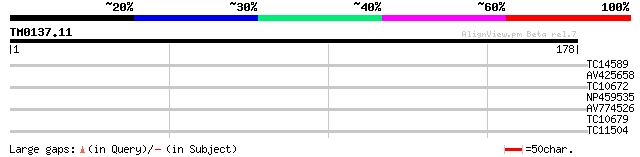
(178 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC14589 similar to UP|Q9SI72 (Q9SI72) F23N19.14, partial (38%) 33 0.037
AV425658 27 1.5
TC10672 weakly similar to PIR|T00710|T00710 thioredoxin homolog ... 27 2.0
NP459535 cytochrome f [Lotus japonicus] 26 3.4
AV774526 26 4.5
TC10679 weakly similar to UP|LOXA_PHAVU (P27480) Lipoxygenase 1 ... 26 4.5
TC11504 similar to UP|O82273 (O82273) At2g31110 protein, partial... 25 5.9
>TC14589 similar to UP|Q9SI72 (Q9SI72) F23N19.14, partial (38%)
Length = 826
Score = 32.7 bits (73), Expect = 0.037
Identities = 27/129 (20%), Positives = 53/129 (40%), Gaps = 10/129 (7%)
Frame = +2
Query: 32 PSDAQLVAKTCKNTPYPSACLQFLQADPRSSSADVTGLALIMVDVIQAKTNGVLNKISQL 91
P+ + + +C+ T YP+ C+Q L + LA+ + V + T + + ++
Sbjct: 146 PNPIEFIKFSCRATRYPAVCVQTLTRYAHVIRQNEQQLAITALTVSMSMTKSSASFMKKM 325
Query: 92 LKGGGDKPALNSCQGRYNAILKADIPQATQALK------TGNPKF----AEDGVADAGVE 141
K G KP + + + + Q++K GN + + V+ A +
Sbjct: 326 TKVKGIKPREHGAVQDCKENMDNSVDRLNQSVKEMGLTAAGNVMWRMSNVQTWVSAALTD 505
Query: 142 ANTCESGFS 150
NTC GF+
Sbjct: 506 QNTCLDGFA 532
>AV425658
Length = 280
Score = 27.3 bits (59), Expect = 1.5
Identities = 18/56 (32%), Positives = 25/56 (44%), Gaps = 3/56 (5%)
Frame = +1
Query: 3 NLKSLSLICNI---FIVVATISMSIGHCRVLQPSDAQLVAKTCKNTPYPSACLQFL 55
N+ S S +CNI F+++ SI P+ CK+TP PS C L
Sbjct: 34 NMASKSTLCNIPLTFLILLPFFASIAFSDT-PPTTPVSPGTACKSTPDPSYCKSVL 198
>TC10672 weakly similar to PIR|T00710|T00710 thioredoxin homolog F22O13.5 -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(12%)
Length = 511
Score = 26.9 bits (58), Expect = 2.0
Identities = 16/56 (28%), Positives = 29/56 (51%), Gaps = 2/56 (3%)
Frame = +3
Query: 9 LICNIFIVVATISMSIGHCRVLQPSDAQLVAKTCKNTPYPSACL--QFLQADPRSS 62
L+ +F + +S +GH + + +V+K K+ +PS L Q L++ P SS
Sbjct: 156 LMAEVFTNTSLVSSWLGHTQQHHHNQKLMVSKVAKSCSFPSLKLKSQALRSSPFSS 323
>NP459535 cytochrome f [Lotus japonicus]
Length = 963
Score = 26.2 bits (56), Expect = 3.4
Identities = 12/33 (36%), Positives = 21/33 (63%)
Frame = +1
Query: 75 DVIQAKTNGVLNKISQLLKGGGDKPALNSCQGR 107
+V A T+G++NKI + KGG + +++ GR
Sbjct: 610 NVYNATTSGIINKIIRKDKGGYEITIVDASDGR 708
>AV774526
Length = 472
Score = 25.8 bits (55), Expect = 4.5
Identities = 8/27 (29%), Positives = 19/27 (69%)
Frame = -1
Query: 7 LSLICNIFIVVATISMSIGHCRVLQPS 33
++ IC ++ ++ T+S+ + CR++ PS
Sbjct: 88 VTCICRLWTILWTLSIDLSGCRLVAPS 8
>TC10679 weakly similar to UP|LOXA_PHAVU (P27480) Lipoxygenase 1 , partial
(22%)
Length = 615
Score = 25.8 bits (55), Expect = 4.5
Identities = 17/60 (28%), Positives = 27/60 (44%), Gaps = 2/60 (3%)
Frame = +3
Query: 75 DVIQAKTNGVLNKISQLLKGGGD--KPALNSCQGRYNAILKADIPQATQALKTGNPKFAE 132
D+I A GVL LL+ GD A+++ ++ + + AT+ GN K E
Sbjct: 120 DIISATKGGVLGLAGDLLEFAGDVAGQAVDTATAIFSRSVAIQLISATKTDANGNGKLGE 299
>TC11504 similar to UP|O82273 (O82273) At2g31110 protein, partial (40%)
Length = 579
Score = 25.4 bits (54), Expect = 5.9
Identities = 15/46 (32%), Positives = 21/46 (45%)
Frame = -3
Query: 32 PSDAQLVAKTCKNTPYPSACLQFLQADPRSSSADVTGLALIMVDVI 77
P Q + YPS C FL + +S TGL LI+V+ +
Sbjct: 190 PGRHQWLQSVAMTPLYPSGCASFLYCESVVTSKKYTGL-LILVNTL 56
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.131 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,782,825
Number of Sequences: 28460
Number of extensions: 31984
Number of successful extensions: 154
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 154
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 154
length of query: 178
length of database: 4,897,600
effective HSP length: 84
effective length of query: 94
effective length of database: 2,506,960
effective search space: 235654240
effective search space used: 235654240
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 52 (24.6 bits)
Lotus: description of TM0137.11


