

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
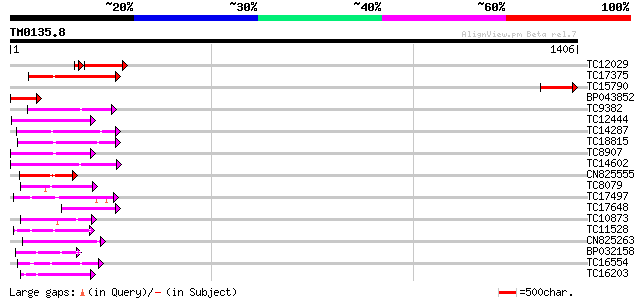
Query= TM0135.8
(1406 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC12029 homologue to UP|Q9STE0 (Q9STE0) MAP3K epsilon 1 protein ... 228 5e-68
TC17375 mitogen-activated kinase kinase kinase alpha [Lotus corn... 172 3e-43
TC15790 similar to UP|Q9STE0 (Q9STE0) MAP3K epsilon 1 protein ki... 170 1e-42
BP043852 142 4e-34
TC9382 weakly similar to UP|ABL_DROME (P00522) Tyrosine-protein ... 138 5e-33
TC12444 similar to UP|O22981 (O22981) Ser/Thr protein kinase iso... 122 3e-28
TC14287 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein k... 115 5e-26
TC18815 similar to UP|Q8W1D5 (Q8W1D5) CBL-interacting protein ki... 110 2e-24
TC8907 similar to UP|Q8H0X3 (Q8H0X3) Serine/threonine protein ki... 108 8e-24
TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinas... 106 2e-23
CN825555 102 5e-22
TC8079 homologue to UP|Q9M6S1 (Q9M6S1) MAP kinase 3, complete 99 5e-21
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 94 1e-19
TC17648 similar to UP|Q9AYN9 (Q9AYN9) NQK1 MAPKK, partial (51%) 94 2e-19
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 92 7e-19
TC11528 homologue to UP|Q39868 (Q39868) Protein kinase , partial... 91 1e-18
CN825263 91 1e-18
BP032158 91 2e-18
TC16554 homologue to UP|AAN71719 (AAN71719) Glycogen synthase ki... 90 3e-18
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 90 3e-18
>TC12029 homologue to UP|Q9STE0 (Q9STE0) MAP3K epsilon 1 protein kinase,
partial (10%)
Length = 874
Score = 228 bits (582), Expect(2) = 5e-68
Identities = 107/108 (99%), Positives = 107/108 (99%)
Frame = +3
Query: 185 WMAPEVIEMSGVCAASDIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQDEQPPIPDSLS 244
WMAPEVIEMSGVCAASDIWSVGCT IELLTCVPPYYDLQPMPALFRIVQDEQPPIPDSLS
Sbjct: 72 WMAPEVIEMSGVCAASDIWSVGCTFIELLTCVPPYYDLQPMPALFRIVQDEQPPIPDSLS 251
Query: 245 PNITDFLHQCFKKDARQRPDAKTLLSHPWILNCRRVLQSSLRHSGTLR 292
PNITDFLHQCFKKDARQRPDAKTLLSHPWILNCRRVLQSSLRHSGTLR
Sbjct: 252 PNITDFLHQCFKKDARQRPDAKTLLSHPWILNCRRVLQSSLRHSGTLR 395
Score = 48.1 bits (113), Expect(2) = 5e-68
Identities = 22/22 (100%), Positives = 22/22 (100%)
Frame = +2
Query: 162 DFGVATKLTEADINTHSVVGTP 183
DFGVATKLTEADINTHSVVGTP
Sbjct: 2 DFGVATKLTEADINTHSVVGTP 67
>TC17375 mitogen-activated kinase kinase kinase alpha [Lotus corniculatus
var. japonicus]
Length = 1422
Score = 172 bits (437), Expect = 3e-43
Identities = 92/233 (39%), Positives = 144/233 (61%), Gaps = 5/233 (2%)
Frame = +3
Query: 47 AIKQVSL---ENIAQEDLNIIMEMINILQNLNHKNIVKYLGSLKTKSHLHIILEYVENGS 103
AIK+V + + ++E L + + IN+L +H NIV+Y GS + L + LEYV GS
Sbjct: 12 AIKEVKVFSDDKTSKECLKQLNQEINLLNQFSHPNIVQYYGSELGEESLSVYLEYVSGGS 191
Query: 104 LANIIKPNKFGPFPESLVAVYIAQVLEGLVYLHEQGVIHRDIKGANILTTKEGLVKLADF 163
+ +++ ++G F E ++ Y Q++ GL YLH + +HRDIKGANIL G +KLADF
Sbjct: 192 IHKLLQ--EYGAFKEPVIQNYTRQIVSGLAYLHSRNTVHRDIKGANILVDPNGEIKLADF 365
Query: 164 GVATKLTEADINTHSVVGTPYWMAPEVI-EMSGVCAASDIWSVGCTVIELLTCVPPYYDL 222
G++ + A + S G+PYWMAPEV+ +G DI S+GCT++E+ T PP+
Sbjct: 366 GMSKHINSA-ASMLSFKGSPYWMAPEVVMNTNGYGLPVDISSLGCTILEMATSKPPWSQF 542
Query: 223 QPMPALFRIVQD-EQPPIPDSLSPNITDFLHQCFKKDARQRPDAKTLLSHPWI 274
+ + A+F+I + P IP+ LS + +F+ QC ++D RP A++LL+HP+I
Sbjct: 543 EGVAAIFKIGNSKDMPEIPEHLSDDAKNFIKQCLQRDPLARPTAQSLLNHPFI 701
>TC15790 similar to UP|Q9STE0 (Q9STE0) MAP3K epsilon 1 protein kinase, partial
(7%)
Length = 579
Score = 170 bits (431), Expect = 1e-42
Identities = 87/90 (96%), Positives = 89/90 (98%)
Frame = +1
Query: 1317 SSRINTTLAVNGLTPLLIARLDHQDAIARLNLLRLIKAVYEHHPQPKKLIVENDLPEKLQ 1376
SSRINTTL+VNGLTPLLIARLDHQDAIARLNLLRLIKAVYEHHPQP+KLIVENDLPEKL
Sbjct: 1 SSRINTTLSVNGLTPLLIARLDHQDAIARLNLLRLIKAVYEHHPQPQKLIVENDLPEKLP 180
Query: 1377 NLIGERRDGQVLVKQMATSLLKALHINTVL 1406
NLIGERRDGQVLVKQMATSLLKALHINTVL
Sbjct: 181 NLIGERRDGQVLVKQMATSLLKALHINTVL 270
>BP043852
Length = 524
Score = 142 bits (358), Expect = 4e-34
Identities = 71/79 (89%), Positives = 75/79 (94%)
Frame = +3
Query: 1 MSRQSTSSAFTKSKTLDNKYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQED 60
MSRQSTSSAFTKSKTLDNKYMLGDE GKGAYGRVYKG DLENGD VAIKQVSLENIAQED
Sbjct: 288 MSRQSTSSAFTKSKTLDNKYMLGDEXGKGAYGRVYKGXDLENGDXVAIKQVSLENIAQED 467
Query: 61 LNIIMEMINILQNLNHKNI 79
LNIIM+ I++L+NLNHKNI
Sbjct: 468 LNIIMQEIDLLKNLNHKNI 524
>TC9382 weakly similar to UP|ABL_DROME (P00522) Tyrosine-protein kinase Abl
(D-ash) , partial (5%)
Length = 1197
Score = 138 bits (348), Expect = 5e-33
Identities = 78/220 (35%), Positives = 128/220 (57%), Gaps = 1/220 (0%)
Frame = +2
Query: 45 FVAIKQVSLENIAQEDLNIIMEMINILQNLNHKNIVKYLGSLKTKSHLHIILEYVENGSL 104
+VAIK + E I+ + L + + I++ + HKN+V+++G+ +L I+ E++ GSL
Sbjct: 14 YVAIKVLKPERISTDMLKEFAQEVYIMRKIRHKNVVQFIGACTRTPNLCIVTEFMSRGSL 193
Query: 105 ANIIKPNKFGPFPESLVAVYIAQVLEGLVYLHEQGVIHRDIKGANILTTKEGLVKLADFG 164
+ + + G F + V +G+ YLH+ +IHRD+K N+L + LVK+ADFG
Sbjct: 194 YDFLHKQR-GVFKLPSLLKVAIDVSKGMNYLHQNNIIHRDLKTGNLLMDENELVKVADFG 370
Query: 165 VATKLTEADINTHSVVGTPYWMAPEVIEMSGVCAASDIWSVGCTVIELLTCVPPYYDLQP 224
VA +T++ + T + GT WMAPEVIE +D++S G + ELLT PY L P
Sbjct: 371 VARVITQSGVMT-AETGTYRWMAPEVIEHKPYDQKADVFSFGIALWELLTGELPYSYLTP 547
Query: 225 MPALFRIVQDE-QPPIPDSLSPNITDFLHQCFKKDARQRP 263
+ A +VQ +P IP + P +++ L +C+K+D +RP
Sbjct: 548 LQAAVGVVQKGLRPSIPKNTHPRLSELLQRCWKQDPIERP 667
>TC12444 similar to UP|O22981 (O22981) Ser/Thr protein kinase isolog,
partial (37%)
Length = 709
Score = 122 bits (307), Expect = 3e-28
Identities = 73/214 (34%), Positives = 119/214 (55%), Gaps = 5/214 (2%)
Frame = +2
Query: 4 QSTSSAFTKSKTLDN-KYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQEDLN 62
+ST + T++ +L++ Y L +E+G GA VY+ + L + VA+K V L+ +L
Sbjct: 38 RSTMGSGTRTYSLNSGDYNLLEEVGYGASATVYRAVFLPRDEEVAVKCVDLDR-CNANLE 214
Query: 63 IIMEMINILQNLNHKNIVKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESLVA 122
I + ++H+N+V+ S L +++ ++ GS +++K F E +
Sbjct: 215 DIRREAQTMSLIDHRNVVRAHWSFVVDRRLWVVMPFMAQGSCLHLMKVAYPDGFEEEAIG 394
Query: 123 VYIAQVLEGLVYLHEQGVIHRDIKGANILTTKEGLVKLADFGVATKLTEA---DINTHSV 179
+ + L+ L YLH G IHRD+K NIL +G VKLADFGV+ + +A N ++
Sbjct: 395 SILKETLKALEYLHRHGHIHRDVKAGNILLGSDGQVKLADFGVSASMFDAGERQRNRNTF 574
Query: 180 VGTPYWMAPEVIEM-SGVCAASDIWSVGCTVIEL 212
VGTP WMAPEV++ +G +D+WS G T +EL
Sbjct: 575 VGTPCWMAPEVLQPGTGYNFKADVWSFGITALEL 676
>TC14287 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein kinase-like
protein (CBL-interacting protein kinase 5), partial
(75%)
Length = 1999
Score = 115 bits (288), Expect = 5e-26
Identities = 76/261 (29%), Positives = 135/261 (51%), Gaps = 5/261 (1%)
Frame = +3
Query: 18 NKYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQEDL-NIIMEMINILQNLNH 76
NKY +G +G+G + +VY G +L + VAIK + E + ++ L I +++++ + H
Sbjct: 426 NKYEMGRVLGQGNFAKVYHGRNLATNENVAIKVIKKEKLKKDRLVKQIKREVSVMRLVRH 605
Query: 77 KNIVKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGLVYLH 136
+IV+ + TK + +++EYV+ G L K NK G E Y Q++ + + H
Sbjct: 606 PHIVELKEVMATKGKIFLVMEYVKGGEL--FTKVNK-GKLNEDDARKYFQQLISAVDFCH 776
Query: 137 EQGVIHRDIKGANILTTKEGLVKLADFGVATKLTEADINTHSVV---GTPYWMAPEVIEM 193
+GV HRD+K N+L + +K++DFG++ L E + +V GTP ++APEV++
Sbjct: 777 SRGVTHRDLKPENLLLDENEDLKVSDFGLSA-LPEQRRDDGMLVTPCGTPAYVAPEVLKK 953
Query: 194 SGVCAA-SDIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQDEQPPIPDSLSPNITDFLH 252
G + +DIWS G + LL+ P+ M ++R + P+ +SP + +
Sbjct: 954 KGYDGSKADIWSCGVILYALLSGYLPFQGENVM-RIYRKAFKAEYEFPEWISPQAKNLIS 1130
Query: 253 QCFKKDARQRPDAKTLLSHPW 273
D +R ++S PW
Sbjct: 1131NLLVADPEKRYSIPEIISDPW 1193
>TC18815 similar to UP|Q8W1D5 (Q8W1D5) CBL-interacting protein kinase
CIPK25, partial (58%)
Length = 979
Score = 110 bits (274), Expect = 2e-24
Identities = 75/261 (28%), Positives = 129/261 (48%), Gaps = 6/261 (2%)
Frame = +2
Query: 19 KYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQEDL-NIIMEMINILQNLNHK 77
KY +G +GKG +VY ++ +G+ VAIK +S I +E + + I I+I++ + H
Sbjct: 35 KYEMGRVLGKGTLAKVYFAKEITSGEGVAIKVMSKARIKKEGMMDQIKREISIMRLVRHP 214
Query: 78 NIVKYLGSLKTKSHLHIILEYVENGSL-ANIIKPNKFGPFPESLVAVYIAQVLEGLVYLH 136
NIV + TK+ + I+EY+ G L A + K G + L Y Q++ + Y H
Sbjct: 215 NIVNLKEVMATKTKIFFIMEYIRGGELFAKVAK----GKLKDDLARRYFQQLISAVDYCH 382
Query: 137 EQGVIHRDIKGANILTTKEGLVKLADFGVA--TKLTEADINTHSVVGTPYWMAPEVIEMS 194
+GV HRD+K N+L + +K++DFG++ + D H+ GTP ++APEV+
Sbjct: 383 SRGVSHRDLKPENLLLDENENLKVSDFGLSGLPEQLRQDGLLHTQCGTPAYVAPEVLRKK 562
Query: 195 GVCA-ASDIWSVGCTVIELLT-CVPPYYDLQPMPALFRIVQDEQPPIPDSLSPNITDFLH 252
G +D WS G + LL C+P + + + ++ V + P SP +
Sbjct: 563 GYDGFKTDTWSCGVILYALLAGCLP--FQHENLMTMYNKVLRAEFQFPPWFSPESKKLIS 736
Query: 253 QCFKKDARQRPDAKTLLSHPW 273
+ D +R +++ W
Sbjct: 737 KILVADPNRRITISSIMRVSW 799
>TC8907 similar to UP|Q8H0X3 (Q8H0X3) Serine/threonine protein kinase-like
protein, partial (44%)
Length = 941
Score = 108 bits (269), Expect = 8e-24
Identities = 67/216 (31%), Positives = 119/216 (55%), Gaps = 5/216 (2%)
Frame = +3
Query: 3 RQSTSSAFTKSKTLDNKYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQEDL- 61
+QS + K L NKY +G +G+G + +VY G ++E + VAIK + E + +E L
Sbjct: 291 QQSRTMRAAKRNILFNKYEIGKILGQGNFAKVYHGRNMETNESVAIKVIKKERLKKERLV 470
Query: 62 NIIMEMINILQNLNHKNIVKYLGSLKTKSHLHIILEYVENGSL-ANIIKPNKFGPFPESL 120
I +++++ + H +IV+ + TK+ + +++EYV+ G L A + K G E
Sbjct: 471 KQIKREVSVMRLVRHPHIVELKEVMATKTKIFMVVEYVKGGELFAKLTK----GKMTEVA 638
Query: 121 VAVYIAQVLEGLVYLHEQGVIHRDIKGANILTTKEGLVKLADFGVAT--KLTEADINTHS 178
Y Q++ + + H +GV HRD+K N+L +K++DFG+++ + +D +
Sbjct: 639 ARKYFQQLISAVDFCHSRGVTHRDLKPENLLLDDNEDLKVSDFGLSSLPEQRRSDGMLLT 818
Query: 179 VVGTPYWMAPEVIEMSGVCAA-SDIWSVGCTVIELL 213
GTP ++APEV++ G + +DIWS G + LL
Sbjct: 819 PCGTPAYVAPEVLKKKGYDGSKADIWSCGVILYALL 926
>TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinase, complete
Length = 1256
Score = 106 bits (265), Expect = 2e-23
Identities = 79/281 (28%), Positives = 129/281 (45%), Gaps = 6/281 (2%)
Frame = +2
Query: 2 SRQSTSSAFTKSKTLDNKYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQE-D 60
+ QS + L ++Y L +EIG+G +G +++ + D A+K + +A D
Sbjct: 14 TEQSRERK*AMCEALKSQYQLCEEIGRGRFGTIFRCFHPISTDTFAVKLIDKSLLADSTD 193
Query: 61 LNIIMEMINILQNLN-HKNIVKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPES 119
+ + + L+ H NI++ + L +++E + +L + I PE
Sbjct: 194 RHCLENEPKYMSLLSPHPNILQIFDVFEDDDVLSMVIELCQPLTLLDRIVAANGTSIPEV 373
Query: 120 LVAVYIAQVLEGLVYLHEQGVIHRDIKGANILTTKEGLVKLADFGVATKLTEADINTHSV 179
A + Q+LE + + H GV HRD+K N+L G +KLADFG A + V
Sbjct: 374 EAAGLMKQLLEAVAHCHRLGVAHRDVKPDNVLFGGGGDLKLADFGSAEWFGDGR-RMSGV 550
Query: 180 VGTPYWMAPEVIEMSGVCAASDIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQDEQPPI 239
VGTPY++APEV+ D+WS G + +L+ PP+Y +F V
Sbjct: 551 VGTPYYVAPEVLMGREYGEKVDVWSCGVILYIMLSGTPPFYG-DSAAEIFEAVIRGNLRF 727
Query: 240 PD----SLSPNITDFLHQCFKKDARQRPDAKTLLSHPWILN 276
P ++SP D L + +D R A+ L HPW L+
Sbjct: 728 PSRIFRNVSPAAKDLLRKMICRDPSNRISAEQALRHPWFLS 850
>CN825555
Length = 714
Score = 102 bits (253), Expect = 5e-22
Identities = 56/146 (38%), Positives = 89/146 (60%), Gaps = 2/146 (1%)
Frame = +1
Query: 24 DEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQEDLNIIMEMINILQNLNHKNIVKYL 83
D+IG+G Y VYK D G VA+K+V +N+ E + + I IL+ L+H N++K
Sbjct: 196 DKIGQGTYSNVYKARDTLTGKIVALKKVRFDNLEPESVKFMAREILILRRLDHPNVLKLE 375
Query: 84 GSLKTKSH--LHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGLVYLHEQGVI 141
G + ++ L+++ EY+ + LA + N F ES V Y+ Q+ GL + H + V+
Sbjct: 376 GLVTSRMSCSLYLVFEYMVH-DLAGLAT-NPAIKFTESQVKCYMHQLFTGLEHCHNRHVL 549
Query: 142 HRDIKGANILTTKEGLVKLADFGVAT 167
HRDIKG+N+L EG++K+ADFG+A+
Sbjct: 550 HRDIKGSNLLIDNEGVLKIADFGLAS 627
>TC8079 homologue to UP|Q9M6S1 (Q9M6S1) MAP kinase 3, complete
Length = 1550
Score = 99.0 bits (245), Expect = 5e-21
Identities = 64/198 (32%), Positives = 106/198 (53%), Gaps = 6/198 (3%)
Frame = +3
Query: 26 IGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQEDLNIIMEMINILQNLNHKNIVKYLGS 85
IG+GAYG V L+ E + VA+K+++ D + I +L++L+H+N++
Sbjct: 309 IGRGAYGIVCSLLNTETNELVAVKKIANAFDNHMDAKRTLREIKLLRHLDHENVIALRDV 488
Query: 86 L-----KTKSHLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGLVYLHEQGV 140
+ + + ++I E ++ L II+ N+ E ++ QVL GL Y+H +
Sbjct: 489 IPPPLRREFTDVYITTELMDT-DLHQIIRSNQ--GLSEEHCQYFLYQVLRGLKYIHSANI 659
Query: 141 IHRDIKGANILTTKEGLVKLADFGVATKLTEADINTHSVVGTPYWMAPEVI-EMSGVCAA 199
IHRD+K +N+L +K+ DFG+A E D T VV T ++ APE++ S +A
Sbjct: 660 IHRDLKPSNLLLNSNCDLKIIDFGLARPTVENDFMTEYVV-TRWYRAPELLLNSSDYTSA 836
Query: 200 SDIWSVGCTVIELLTCVP 217
D+WSVGC +EL+ P
Sbjct: 837 IDVWSVGCIFMELMNKKP 890
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 94.4 bits (233), Expect = 1e-19
Identities = 75/284 (26%), Positives = 138/284 (48%), Gaps = 24/284 (8%)
Frame = +1
Query: 10 FTKSKTLDNKYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQEDLNIIMEMIN 69
F+ + N + L +++G+G +G VYKGL L NG +A+K++S N + + + I
Sbjct: 1642 FSTISSATNHFSLSNKLGEGGFGPVYKGL-LANGQEIAVKRLS--NTSGQGMEEFKNEIK 1812
Query: 70 ILQNLNHKNIVKYLGSL---KTKSHLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIA 126
++ L H+N+VK G SH + ++ + + + + ++ NK I
Sbjct: 1813 LIARLQHRNLVKLFGCSVHQDENSHANKKMKILLDSTRSKLVDWNKR--------LQIID 1968
Query: 127 QVLEGLVYLHEQG---VIHRDIKGANILTTKEGLVKLADFGVATKL--TEADINTHSVVG 181
+ GL+YLH+ +IHRD+K +NIL E K++DFG+A + + T V+G
Sbjct: 1969 GIARGLLYLHQDSRLRIIHRDLKTSNILLDDEMNPKISDFGLARIFIGDQVEARTKRVMG 2148
Query: 182 TPYWMAPEVIEMSGVCAASDIWSVGCTVIELLT------CVPPYYDLQPMPALFRIVQDE 235
T +M PE SD++S G V+E+++ P++ L + +R+ +E
Sbjct: 2149 TYGYMPPEYAVHGSFSIKSDVFSFGVIVLEIISGKKIGRFYDPHHHLNLLSHAWRLWIEE 2328
Query: 236 QP------PIPDSLSP-NITDFLH---QCFKKDARQRPDAKTLL 269
+P + D + P I ++H C ++ RPD +++
Sbjct: 2329 RPLELVDELLDDPVIPTEILRYIHVALLCVQRRPENRPDMLSIV 2460
>TC17648 similar to UP|Q9AYN9 (Q9AYN9) NQK1 MAPKK, partial (51%)
Length = 741
Score = 93.6 bits (231), Expect = 2e-19
Identities = 60/155 (38%), Positives = 83/155 (52%), Gaps = 9/155 (5%)
Frame = +3
Query: 129 LEGLVYLH-EQGVIHRDIKGANILTTKEGLVKLADFGVATKLTEADINTHSVVGTPYWMA 187
L+GLVYLH E+ VIHRD K +++L +G VK+ DFGV+ L + + VGT +M+
Sbjct: 3 LQGLVYLHNERHVIHRDSKPSHLLVNHQGEVKITDFGVSAMLATSMGQRDTFVGTYNYMS 182
Query: 188 PEVIEMSGVCAASDIWSVGCTVIELLTCVPPYY---DLQPMPALFR----IVQDEQPPIP 240
P I S +SDIWS+G V+E PY D Q P+ + IV+ P P
Sbjct: 183 PARISGSTYDYSSDIWSLGMVVLECAIGRFPYIQSEDQQAWPSFYELLAAIVESPPPSAP 362
Query: 241 -DSLSPNITDFLHQCFKKDARQRPDAKTLLSHPWI 274
D SP F+ C +KD + R + LL HP+I
Sbjct: 363 SDQFSPEFCSFVSSCIQKDPQDRLTSLELLDHPFI 467
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 91.7 bits (226), Expect = 7e-19
Identities = 73/197 (37%), Positives = 105/197 (53%), Gaps = 8/197 (4%)
Frame = +1
Query: 26 IGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQEDLNIIMEMINILQNLNHKNIVKYLGS 85
IG+G +G+VYKG L G+ VA+KQ+S + Q +ME++ +L L+H N+V+ +G
Sbjct: 469 IGEGGFGKVYKGR-LTTGEAVAVKQLSHDG-RQGFQEFVMEVL-MLSLLHHTNLVRLIGY 639
Query: 86 LKTKSHLHIILEYVENGSLANIIKPNKFGPFP-----ESLVAVYIAQVLEGLVYLHEQGV 140
++ EY+ GSL + + P VAV A+ LE L + V
Sbjct: 640 CTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAARGLEYLHCTADPPV 819
Query: 141 IHRDIKGANILTTKEGLVKLADFGVATKLTEADINTH---SVVGTPYWMAPEVIEMSGVC 197
I+RD+K ANIL E KL+DFG+A KL NTH V+GT + APE +
Sbjct: 820 IYRDLKSANILLDNEFNPKLSDFGLA-KLGPVGDNTHVSTRVMGTYGYCAPEYAMSGKLT 996
Query: 198 AASDIWSVGCTVIELLT 214
SDI+S G ++ELLT
Sbjct: 997 LKSDIYSFGVVLLELLT 1047
>TC11528 homologue to UP|Q39868 (Q39868) Protein kinase , partial (56%)
Length = 640
Score = 91.3 bits (225), Expect = 1e-18
Identities = 69/206 (33%), Positives = 102/206 (49%), Gaps = 5/206 (2%)
Frame = +3
Query: 9 AFTKSKTLDNKYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQEDLNIIMEMI 68
AF+K ++ +Y E+G G +G D G+ VA+K + E + D N+ E+I
Sbjct: 48 AFSKVD-MEERYEPLKELGSGNFGVARLARDKNTGELVAVKYI--ERGKKIDENVQREII 218
Query: 69 NILQNLNHKNIVKYLGSLKTKSHLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQV 128
N ++L H NI+++ L T +HL I+LEY G L I G F E + Q+
Sbjct: 219 NH-RSLRHPNIIRFKEVLLTPTHLAIVLEYASGGELFERI--CSAGRFSEDEARYFFQQL 389
Query: 129 LEGLVYLHEQGVIHRDIKGANILT--TKEGLVKLADFGVATKLTEADINTHSVVGTPYWM 186
+ G+ Y H + HRD+K N L +K+ DFG +K S VGTP ++
Sbjct: 390 ISGVSYCHSMEICHRDLKLENTLLDGNPSPRLKICDFGY-SKSAILHSQPKSTVGTPAYI 566
Query: 187 APEVI---EMSGVCAASDIWSVGCTV 209
APEV+ E G A D+WS G T+
Sbjct: 567 APEVLSRKEYDGKVA--DVWSCGVTL 638
>CN825263
Length = 663
Score = 90.9 bits (224), Expect = 1e-18
Identities = 73/215 (33%), Positives = 110/215 (50%), Gaps = 8/215 (3%)
Frame = +1
Query: 31 YGRVYKGLDLENGDFVAIKQVSLENIAQEDLNIIMEMINILQNLNHKNIVKYLGSLKTKS 90
+G VYKG+ L +G VA+K + ++ Q + + +L L+H+N+VK +G K
Sbjct: 4 FGLVYKGI-LNDGRDVAVKILKRDD--QRGGREFLAEVEMLSRLHHRNLVKLIGICIEKQ 174
Query: 91 HLHIILEYVENGSLANIIK--PNKFGPFPESLVAVYIAQVLEGLVYLHEQG---VIHRDI 145
+I E V NGS+ + + + GP + GL YLHE VIHRD
Sbjct: 175 TRCLIYELVPNGSVESHLHGADKETGPLDWNARMKIALGAARGLAYLHEDSNPCVIHRDF 354
Query: 146 KGANILTTKEGLVKLADFGVA-TKLTEAD--INTHSVVGTPYWMAPEVIEMSGVCAASDI 202
K +NIL + K++DFG+A T L E + I+TH V+GT ++APE + SD+
Sbjct: 355 KSSNILLECDFTPKVSDFGLARTALDEGNKHISTH-VMGTFGYLAPEYAMTGHLLVKSDV 531
Query: 203 WSVGCTVIELLTCVPPYYDLQPMPALFRIVQDEQP 237
+S G ++ELLT P DL P +V +P
Sbjct: 532 YSYGVVLLELLTGTKP-VDLSQPPGQENLVTWARP 633
>BP032158
Length = 555
Score = 90.5 bits (223), Expect = 2e-18
Identities = 62/170 (36%), Positives = 97/170 (56%), Gaps = 6/170 (3%)
Frame = +2
Query: 14 KTLDNKYMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQEDLNIIMEMINILQN 73
K N + ++IG+G +G VYKG+ L GD +A+KQ+S ++ Q + I E I ++
Sbjct: 32 KAATNNFDPANKIGEGGFGPVYKGV-LSEGDVIAVKQLSSKS-KQGNREFINE-IGMISA 202
Query: 74 LNHKNIVKYLGSLKTKSHLHIILEYVENGSLANIIKPN---KFGPFPESLVAVYIAQVLE 130
L H N+VK G + L ++ EY+EN SLA + N K + + + + + +
Sbjct: 203 LQHPNLVKLYGCCIEGNQLLLVYEYMENNSLARALFGNEEQKLNLNWRTRMKICVG-IAK 379
Query: 131 GLVYLHEQG---VIHRDIKGANILTTKEGLVKLADFGVATKLTEADINTH 177
GL YLHE+ ++HRDIK N+L K+ K++DFG+A KL E + NTH
Sbjct: 380 GLAYLHEESRLKIVHRDIKATNVLLDKDLKAKISDFGLA-KLDEEE-NTH 523
>TC16554 homologue to UP|AAN71719 (AAN71719) Glycogen synthase kinase 3 beta
protein kinase DWARF12 , partial (71%)
Length = 927
Score = 89.7 bits (221), Expect = 3e-18
Identities = 71/224 (31%), Positives = 114/224 (50%), Gaps = 10/224 (4%)
Frame = +1
Query: 20 YMLGDEIGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQEDLNIIMEMINILQNLNHKNI 79
YM +G G++G V++ LE+G+ VAIK+V +D + +++ ++H N+
Sbjct: 238 YMAERVVGTGSFGIVFQAKCLESGEAVAIKKVL------QDRRYKNRELQLMRVMDHPNV 399
Query: 80 VK----YLGSLKTKS-HLHIILEYVENGSLANIIK--PNKFGPFPESLVAVYIAQVLEGL 132
V + + T L++++EYV S+ +IK N P V +Y+ Q+ GL
Sbjct: 400 VSLKHCFFSTTSTDELFLNLVMEYVPE-SMYRVIKHYSNANQRMPIIYVKLYMYQIFRGL 576
Query: 133 VYLHE-QGVIHRDIKGANILTTK-EGLVKLADFGVATKLTEADINTHSVVGTPYWMAPEV 190
Y+H GV HRD+K NIL VKL DFG A L + + N S + + ++ APE+
Sbjct: 577 AYIHTVPGVCHRDLKPQNILVDPLTHQVKLCDFGSAKMLVKGEANI-SYICSRFYRAPEL 753
Query: 191 IEMSGVCAAS-DIWSVGCTVIELLTCVPPYYDLQPMPALFRIVQ 233
I + + S DIWS GC + ELL P + + L I++
Sbjct: 754 IFGATEYSTSIDIWSAGCVLAELLLGQPLFPGENAVDQLVHIIK 885
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 89.7 bits (221), Expect = 3e-18
Identities = 63/193 (32%), Positives = 97/193 (49%), Gaps = 5/193 (2%)
Frame = +2
Query: 26 IGKGAYGRVYKGLDLENGDFVAIKQVSLENIAQEDLNIIMEMINILQNLNHKNIVKYLGS 85
IGKG G VY+G + NG VAIK++ + + D E I L + H+NI++ LG
Sbjct: 2210 IGKGGAGIVYRG-SMPNGTDVAIKRLVGQGSGRNDYGFRAE-IETLGKIRHRNIMRLLGY 2383
Query: 86 LKTKSHLHIILEYVENGSLANIIKPNKFGPFPESLVAVYIAQVLEGLVYLHEQG---VIH 142
+ K ++ EY+ NGSL + K G + + GL Y+H +IH
Sbjct: 2384 VSNKDTNLLLYEYMPNGSLGEWLHGAKGGHLRWEMRYKIAVEAARGLCYMHHDCSPLIIH 2563
Query: 143 RDIKGANILTTKEGLVKLADFGVATKLTE--ADINTHSVVGTPYWMAPEVIEMSGVCAAS 200
RD+K NIL + +ADFG+A L + A + S+ G+ ++APE V S
Sbjct: 2564 RDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEKS 2743
Query: 201 DIWSVGCTVIELL 213
D++S G ++EL+
Sbjct: 2744 DVYSFGVVLLELI 2782
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.315 0.132 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,514,649
Number of Sequences: 28460
Number of extensions: 293301
Number of successful extensions: 1910
Number of sequences better than 10.0: 241
Number of HSP's better than 10.0 without gapping: 1752
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1779
length of query: 1406
length of database: 4,897,600
effective HSP length: 102
effective length of query: 1304
effective length of database: 1,994,680
effective search space: 2601062720
effective search space used: 2601062720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Lotus: description of TM0135.8


