

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
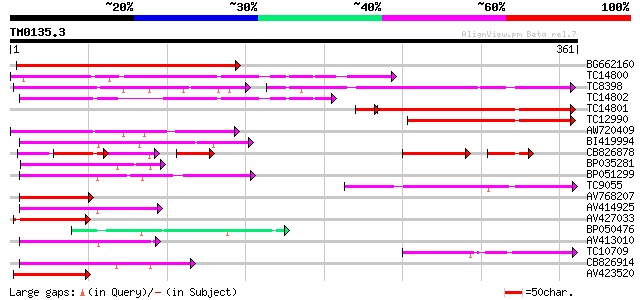
Query= TM0135.3
(361 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BG662160 296 3e-81
TC14800 weakly similar to UP|Q9FHP3 (Q9FHP3) Genomic DNA, chromo... 196 5e-51
TC8398 weakly similar to UP|Q84KK1 (Q84KK1) S haplotype-specific... 108 6e-44
TC14802 weakly similar to UP|CAD56661 (CAD56661) S locus F-box (... 149 9e-37
TC14801 127 9e-31
TC12990 113 4e-26
AW720409 97 3e-21
BI419994 77 6e-15
CB826878 67 1e-14
BP035281 74 4e-14
BP051299 70 6e-13
TC9055 65 2e-11
AV768207 59 1e-09
AV414925 58 2e-09
AV427033 58 3e-09
BP050476 55 1e-08
AV413010 55 2e-08
TC10709 54 3e-08
CB826914 54 5e-08
AV423520 52 1e-07
>BG662160
Length = 445
Score = 296 bits (759), Expect = 3e-81
Identities = 143/143 (100%), Positives = 143/143 (100%)
Frame = +2
Query: 5 QFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFAYLQ 64
QFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFAYLQ
Sbjct: 17 QFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFAYLQ 196
Query: 65 SLITSPRKSRSASTIAIPDDYDFCGTCNGLVCLHSSKYDRKDKYTSHVRLWNPAMRSMSQ 124
SLITSPRKSRSASTIAIPDDYDFCGTCNGLVCLHSSKYDRKDKYTSHVRLWNPAMRSMSQ
Sbjct: 197 SLITSPRKSRSASTIAIPDDYDFCGTCNGLVCLHSSKYDRKDKYTSHVRLWNPAMRSMSQ 376
Query: 125 RSPPLYLPIVYDSYFGFDYDSST 147
RSPPLYLPIVYDSYFGFDYDSST
Sbjct: 377 RSPPLYLPIVYDSYFGFDYDSST 445
>TC14800 weakly similar to UP|Q9FHP3 (Q9FHP3) Genomic DNA, chromosome 5, TAC
clone:K14B20, partial (7%)
Length = 782
Score = 196 bits (498), Expect = 5e-51
Identities = 131/272 (48%), Positives = 159/272 (58%), Gaps = 26/272 (9%)
Frame = +3
Query: 1 MEPPQFL--PEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLH-LHRSSSTTRN 57
MEP L P+EL +EILSWLPVKSL++FR VSK+WKS ISD QF+KLH LHR S RN
Sbjct: 24 MEPLHLLCLPDELIIEILSWLPVKSLLQFRVVSKTWKSFISDPQFVKLHLLHRLS--FRN 197
Query: 58 TDFAY---------------------LQSLITSPRKSRSASTIAIPDDYDFCGTCNGLVC 96
DF + + SL+ SP S ++ + YDF GTCNGLV
Sbjct: 198 ADFEHTSLLIKCHTDDFGRPYISSRTVSSLLESP--SAIVASRSCISGYDFIGTCNGLVS 371
Query: 97 LHSSKYDRKDKYT-SHVRLWNPAMRSMSQRSPPLYLPIVYDSYFGFDYDSSTDTYKVVAV 155
L YD + S VR WNPA R+MSQ SPP + P + + GF YD S+DTYKVV +
Sbjct: 372 LRKLNYDESNTNNFSQVRFWNPATRTMSQDSPPSWSP--RNLHLGFGYDCSSDTYKVVGM 545
Query: 156 AVALSWDTLETMVNVYNIGDDTCWRTIPVSHLPSMYLKDDDAVYVNNTLNRLAILPNVED 215
L TMVNVYN+GD+ CWRTI +S M+L+ AVYV+NTLN LA
Sbjct: 546 IPGL------TMVNVYNMGDN-CWRTIQISPHAPMHLQ-GSAVYVSNTLNWLA------G 683
Query: 216 RDRCTILSFDFGKESCA-QLPLPYCPQSRYDF 246
+ I+SFD KE CA QL LPYCP R +F
Sbjct: 684 TNPYFIVSFDLEKEECAQQLSLPYCPWRRKEF 779
>TC8398 weakly similar to UP|Q84KK1 (Q84KK1) S haplotype-specific F-box
protein a, partial (9%)
Length = 1602
Score = 108 bits (269), Expect(2) = 6e-44
Identities = 75/206 (36%), Positives = 106/206 (51%), Gaps = 9/206 (4%)
Frame = +1
Query: 164 LETMVNVYNIGDDTCWRTIPV------SHLPSMYLKDDDAVYVNNTLNRLAILPNVEDRD 217
++++ NVYN+GD CWR I H D + VY+N T+N + L D
Sbjct: 721 MQSVANVYNMGDG-CWRRIQSFPDFTREHWGRGTAPDQNGVYLNGTINWVLRLNIAYD-- 891
Query: 218 RCTILSFDFGKESCAQLPLP-YCPQSRYDFYRTWPTLGVLRDCLCICQKDKNMHFVVWQM 276
I+S D G E C++L LP + P Y P LGVL+D LC+ + + FV+W+M
Sbjct: 892 ---IISLDLGSEVCSRLSLPCFPPPGPKCVYDLPPILGVLKDSLCVLHNNNDKTFVIWKM 1062
Query: 277 KEFGLYKSWTLLFNIAGYAYDIPCRFFAMYMSENGDALLLSKLAVSQPQAVLYTQKDNKL 336
EFG ++SWT LFN D+ RF + SEN + LL+ + + VLY D
Sbjct: 1063NEFGAHESWTPLFNFDFKKEDL-YRFDKFFFSENDEVLLM-----TLTKQVLYNHSDITF 1224
Query: 337 EVTD-VANYIFNCYAM-NYIESLVPP 360
+ D V NYI + + NY+ESLV P
Sbjct: 1225KFNDRVHNYIHDWNNIENYVESLVSP 1302
Score = 86.3 bits (212), Expect(2) = 6e-44
Identities = 76/208 (36%), Positives = 92/208 (43%), Gaps = 57/208 (27%)
Frame = +3
Query: 3 PPQFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFAY 62
PP L +EL EILS LPVKSLVRFRCV KSWK IS+ QF+KLHL RSS+ DFA
Sbjct: 66 PPPILIDELIWEILSLLPVKSLVRFRCVCKSWKLTISNPQFMKLHLRRSSAKAA-ADFAD 242
Query: 63 LQSLITSPR----KSRSASTIAIPDDYDF------------------------------- 87
Q ++ + R S S I D F
Sbjct: 243 SQVVVMTKRDHIAHSPLLSKFHIVHDETFTADEPWPVHALREPFNQPHVSICYAPSLFSS 422
Query: 88 ------------CGTCNGLVCLHSSKYDRKDKYT--SHVRLWNPAMRSMSQRSPPLYL-- 131
G CNGLV + + Y+ + T S L NPA + SQ SP L L
Sbjct: 423 QNNDKLKGHCRLIGACNGLVSVINEGYNSAESATKLSCCNL-NPATKLRSQVSPTLSLCY 599
Query: 132 ----PIVYDSY--FGFDYDSSTDTYKVV 153
+++ + FGF YD DTYKVV
Sbjct: 600 KQFHSLIHSVFRIFGFGYDPLNDTYKVV 683
>TC14802 weakly similar to UP|CAD56661 (CAD56661) S locus F-box (SLF)-S4
protein, partial (10%)
Length = 551
Score = 149 bits (375), Expect = 9e-37
Identities = 96/203 (47%), Positives = 118/203 (57%), Gaps = 1/203 (0%)
Frame = +3
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHL-HRSSSTTRNTDFAYLQS 65
LP+EL +EILSWLPVKSL++FR VSK+WKS ISD QF+KLHL HR S RN DF +
Sbjct: 21 LPDELIIEILSWLPVKSLLQFRVVSKTWKSFISDPQFVKLHLLHRLSF--RNADFEHTSL 194
Query: 66 LITSPRKSRSASTIAIPDDYDFCGTCNGLVCLHSSKYDRKDKYTSHVRLWNPAMRSMSQR 125
LI H+ + R + VR WNPA R+MSQ
Sbjct: 195 LIKC----------------------------HTDDFGRPYISSRTVRFWNPATRTMSQD 290
Query: 126 SPPLYLPIVYDSYFGFDYDSSTDTYKVVAVAVALSWDTLETMVNVYNIGDDTCWRTIPVS 185
SPP + P + + GF YD S+DTYKVV + L TMVNVYN+GD+ CWRTI +S
Sbjct: 291 SPPSWSP--RNLHLGFGYDCSSDTYKVVGMIPGL------TMVNVYNMGDN-CWRTIQIS 443
Query: 186 HLPSMYLKDDDAVYVNNTLNRLA 208
M+L+ AVYV+NTLN LA
Sbjct: 444 PHAPMHLQ-GSAVYVSNTLNWLA 509
>TC14801
Length = 595
Score = 127 bits (319), Expect(2) = 9e-31
Identities = 71/127 (55%), Positives = 82/127 (63%), Gaps = 1/127 (0%)
Frame = +2
Query: 235 PLPYCPQSRYDFYRTWPTLGVLRDCLCICQKDKNMH-FVVWQMKEFGLYKSWTLLFNIAG 293
PLPYCP R +F LGVLR CLCI Q K H FVV QMKE+G+ +SWT LFNI
Sbjct: 47 PLPYCPWRRQEFDHFGSILGVLRGCLCISQYHKETHSFVVSQMKEYGVNESWTQLFNIIV 226
Query: 294 YAYDIPCRFFAMYMSENGDALLLSKLAVSQPQAVLYTQKDNKLEVTDVANYIFNCYAMNY 353
+ + FAM MSENGD LL ++ VS QAVLY Q+DNKLE T +AN I Y NY
Sbjct: 227 HERILHSPCFAMCMSENGDVLLFAESGVS--QAVLYNQRDNKLEDTKIANNIVGSYVRNY 400
Query: 354 IESLVPP 360
IESL+ P
Sbjct: 401 IESLISP 421
Score = 22.7 bits (47), Expect(2) = 9e-31
Identities = 9/15 (60%), Positives = 11/15 (73%)
Frame = +1
Query: 221 ILSFDFGKESCAQLP 235
I+SFD KE CA+ P
Sbjct: 1 IVSFDLEKEECARPP 45
>TC12990
Length = 518
Score = 113 bits (283), Expect = 4e-26
Identities = 62/108 (57%), Positives = 72/108 (66%), Gaps = 1/108 (0%)
Frame = +1
Query: 254 GVLRDCLCICQKDKNMH-FVVWQMKEFGLYKSWTLLFNIAGYAYDIPCRFFAMYMSENGD 312
GVL CLCI Q +K H FVVWQMKEFG+++SWT LFNI + + FAM MSENGD
Sbjct: 1 GVLGGCLCISQNNKETHSFVVWQMKEFGVHESWTQLFNIIVHERILHTPCFAMCMSENGD 180
Query: 313 ALLLSKLAVSQPQAVLYTQKDNKLEVTDVANYIFNCYAMNYIESLVPP 360
ALL +K VS QAVLY Q++ K E T + N I Y NYIESLV P
Sbjct: 181 ALLFAKSGVS--QAVLYNQREKKFEDTKIGNNIVGSYVRNYIESLVSP 318
>AW720409
Length = 575
Score = 97.4 bits (241), Expect = 3e-21
Identities = 70/172 (40%), Positives = 86/172 (49%), Gaps = 26/172 (15%)
Frame = +3
Query: 1 MEPPQFLPEELRLEILSWLPVKSLVRFRCVSKSWKSII-SDSQFIKLHLHRSSSTTRNTD 59
ME LP EL +EILSWLPVK+L+RF CVSKSWKS+I D F KLHL RS N
Sbjct: 75 MEIEFVLPFELWIEILSWLPVKTLMRFSCVSKSWKSLIYQDRDFKKLHLDRSPK-NNNQV 251
Query: 60 FAYLQSLITSPR-----------KSRSASTIAIPDD--------------YDFCGTCNGL 94
LQ + S R + + S+ +I DD +D G+CNGL
Sbjct: 252 ILTLQKPLFSGRTFFPFPVRRLLQDQEPSSSSIIDDIPFEEEDEDEEDNYHDTIGSCNGL 431
Query: 95 VCLHSSKYDRKDKYTSHVRLWNPAMRSMSQRSPPLYLPIVYDSYFGFDYDSS 146
VCL+ + + RLWNPA R +SPPL I +F F YD S
Sbjct: 432 VCLYGI-----NDHGLWFRLWNPATRFRFHKSPPLDAVIGSTLHFAFGYDHS 572
>BI419994
Length = 549
Score = 76.6 bits (187), Expect = 6e-15
Identities = 56/165 (33%), Positives = 86/165 (51%), Gaps = 16/165 (9%)
Frame = +2
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTT-----RNTDFA 61
LP+EL L+IL LPV+SL+RF+ V KSW+ +ISD QF K H + ++S T NT+
Sbjct: 47 LPDELILQILLRLPVRSLLRFKTVCKSWRYLISDPQFTKSHFNLAASPTHRIFLNNTNGF 226
Query: 62 YLQSLITSPRKSRSASTIAI--------PDDYDFCGTCNGLVCLHSSK-YDRKDKYTSHV 112
++SL T +S + + PD++ F + GL L S + + + +V
Sbjct: 227 *IESLDTDSSLHDESSLVHLKFPLAPPHPDNHRFGSSVTGLKVLGSCRGFVLLANHQGNV 406
Query: 113 RLWNPAMRSMSQRSPP--LYLPIVYDSYFGFDYDSSTDTYKVVAV 155
+WNP+ + +R P YL + +G YD S D Y VV +
Sbjct: 407 VVWNPS-TGVRRRVPEWCSYLASQCEFLYGIGYDESNDDYLVVFI 538
>CB826878
Length = 635
Score = 66.6 bits (161), Expect(2) = 1e-14
Identities = 31/44 (70%), Positives = 35/44 (79%), Gaps = 1/44 (2%)
Frame = +3
Query: 251 PTLGVLRDCLCICQKDKNM-HFVVWQMKEFGLYKSWTLLFNIAG 293
P LGVLRD LC+ Q DK +FVVWQMKEFG++KSWT LFNI G
Sbjct: 384 PILGVLRDSLCVSQNDKKTRNFVVWQMKEFGVHKSWTQLFNIIG 515
Score = 42.4 bits (98), Expect = 1e-04
Identities = 21/35 (60%), Positives = 27/35 (77%)
Frame = +3
Query: 29 CVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFAYL 63
C ++S KS+ISDS F+KLHLHR SS RN DF+ +
Sbjct: 105 CQNQSCKSLISDSAFVKLHLHR*SS--RNADFSMI 203
Score = 36.2 bits (82), Expect(2) = 5e-07
Identities = 30/93 (32%), Positives = 44/93 (47%), Gaps = 3/93 (3%)
Frame = +1
Query: 6 FLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFAYLQS 65
FLPEEL +EI SWLPV+S V CV + H+ SS + +F + +
Sbjct: 43 FLPEELVVEIFSWLPVQSHV---CVKIN-------------HVSLSSPILHS*NFTFTGN 174
Query: 66 LITSPRKSRSASTIAIPDDYDF---CGTCNGLV 95
L+ + T+ + +F GTCNGL+
Sbjct: 175 LLEMQIFR*FSPTLYLISLCNFTLGIGTCNGLI 273
Score = 33.5 bits (75), Expect(2) = 5e-07
Identities = 14/24 (58%), Positives = 17/24 (70%)
Frame = +2
Query: 107 KYTSHVRLWNPAMRSMSQRSPPLY 130
KY S + WNPA R+M Q +PPLY
Sbjct: 311 KYCSLLCFWNPATRTMPQFAPPLY 382
Score = 29.3 bits (64), Expect(2) = 1e-14
Identities = 16/29 (55%), Positives = 20/29 (68%)
Frame = +2
Query: 305 MYMSENGDALLLSKLAVSQPQAVLYTQKD 333
+ +SENGDALLLS S QA+LY +D
Sbjct: 554 LILSENGDALLLSTYGGS--QAILYIGRD 634
>BP035281
Length = 498
Score = 73.9 bits (180), Expect = 4e-14
Identities = 46/108 (42%), Positives = 60/108 (54%), Gaps = 16/108 (14%)
Frame = +2
Query: 8 PEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQ-FIKLHLHRSSSTTRNTDFAYLQSL 66
P EL +EILSWLPV SL+RF+CV KSWKS+IS ++ F KL L R+S + S
Sbjct: 125 PSELVIEILSWLPVLSLIRFKCVCKSWKSLISHNKAFAKLQLERTSLKINHVILTSSDSS 304
Query: 67 I---TSPRKSRSASTIAIPDDYDFC------------GTCNGLVCLHS 99
+ PR S+I + D D C G+CNGL+CLH+
Sbjct: 305 FIPCSLPRLLEDPSSI-MDQDQDRCLRLDKFEYNEVIGSCNGLICLHA 445
>BP051299
Length = 539
Score = 70.1 bits (170), Expect = 6e-13
Identities = 49/162 (30%), Positives = 81/162 (49%), Gaps = 12/162 (7%)
Frame = +2
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSST--------TRNT 58
LP+E+ EIL LP K+LV+ V KSW+S+I+ + FI LH + S S+
Sbjct: 5 LPQEIWAEILHRLPPKTLVKCTSVCKSWRSLITSTSFISLHRNHSPSSLLLQLCDERAIP 184
Query: 59 DFAYLQSLITSPRKSRSASTIAIPD----DYDFCGTCNGLVCLHSSKYDRKDKYTSHVRL 114
+F + P S S S++ +P ++ G C+GLVC+ S++ R + +
Sbjct: 185 NFIHYSLRRDDPFLSES-SSLRLPSSFNREFSVVGICHGLVCITSTENCR------DLII 343
Query: 115 WNPAMRSMSQRSPPLYLPIVYDSYFGFDYDSSTDTYKVVAVA 156
NP++R P P ++ + F +DS + YKV+ +A
Sbjct: 344 CNPSLRLHITLPEPSDYPSLHSASVAFGFDSRNNDYKVIRIA 469
>TC9055
Length = 577
Score = 64.7 bits (156), Expect = 2e-11
Identities = 45/152 (29%), Positives = 72/152 (46%), Gaps = 4/152 (2%)
Frame = +2
Query: 214 EDRDRCTILSFDFGKESCAQLPLPYCPQSRYDFYRTWPTLGVLRDCLCICQKDKNMHFVV 273
E ++ I+S D +E+ L LP + VL +CLC+ K HFV
Sbjct: 32 EPFEQLVIVSLDMRQEAYRLLSLPQGTSELPS-----AEIQVLGNCLCLFYDFKRTHFVA 196
Query: 274 WQMKEFGLYKSWTLLFNIAGYAYDIPCRFF----AMYMSENGDALLLSKLAVSQPQAVLY 329
W+M E+GL +SWT L I+ RF+ + + +G +L+K Q ++Y
Sbjct: 197 WKMSEYGLPESWTPLLTISYEHLRWDHRFYLNQWLICLCGDGHIFMLAK---KQKLVIIY 367
Query: 330 TQKDNKLEVTDVANYIFNCYAMNYIESLVPPC 361
DN ++ ++A+ A +Y+ESLV PC
Sbjct: 368 NLSDNSVKYVELASNKLWLDANDYVESLVSPC 463
>AV768207
Length = 374
Score = 59.3 bits (142), Expect = 1e-09
Identities = 30/47 (63%), Positives = 36/47 (75%)
Frame = +1
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSS 53
LP EL +EIL LPVKSL++ RCV KSWKS+ISD +F K HL SS+
Sbjct: 73 LPFELGVEILCRLPVKSLLQLRCVCKSWKSLISDPKFAKNHLRCSST 213
>AV414925
Length = 414
Score = 58.2 bits (139), Expect = 2e-09
Identities = 33/101 (32%), Positives = 52/101 (50%), Gaps = 10/101 (9%)
Frame = +1
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSST----------TR 56
+P E+ +ILS LPV SL+RFR +SKSW+S+I F+KLHL+ S +
Sbjct: 97 IPVEVIADILSRLPVTSLLRFRSISKSWRSLIDSKHFMKLHLNNSLTNPNPNLTNLILRH 276
Query: 57 NTDFAYLQSLITSPRKSRSASTIAIPDDYDFCGTCNGLVCL 97
NTD + + + + + G+C+GL+C+
Sbjct: 277 NTDLYRADFPSIGAAVNLNHPLMCYSNRINILGSCHGLLCI 399
>AV427033
Length = 420
Score = 57.8 bits (138), Expect = 3e-09
Identities = 30/49 (61%), Positives = 36/49 (73%)
Frame = +1
Query: 3 PPQFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRS 51
PP LP +L +EIL LPV SL++FRCV KSW S+ISD +F K HLH S
Sbjct: 118 PP--LPFDLVVEILCRLPVNSLLQFRCVCKSWNSLISDPKFAKNHLHCS 258
>BP050476
Length = 520
Score = 55.5 bits (132), Expect = 1e-08
Identities = 50/169 (29%), Positives = 68/169 (39%), Gaps = 30/169 (17%)
Frame = +2
Query: 40 DSQFIKLHLHRSSSTTRNTDFAYLQSLITSPRKSRSASTIAIP----------------- 82
DS F+KLHL+RS T L ++ P + T +P
Sbjct: 17 DSSFVKLHLNRSPKNTH-----ILLNIANDPYDFENDDTWVVPSSVCCLIEDLSSMIDAK 181
Query: 83 ------DDYDFCGTCNGLVCLHSSKYDRKDKYTSHVRLWNPAMRSMSQRSPPLYLPIVYD 136
D + G+ NGL+C + YD V+LWNPA S++SP L +
Sbjct: 182 GCYLLKDGHLVIGSSNGLICF-GNFYDVGPIEEFWVQLWNPATHLKSKKSPTFNLSMRTS 358
Query: 137 S-------YFGFDYDSSTDTYKVVAVAVALSWDTLETMVNVYNIGDDTC 178
GF YD+ DTYKVV V + +ETM VY + D C
Sbjct: 359 VDAPPGKVNLGFGYDNLHDTYKVVLVHWNCTKQKMETM--VYCVNDIFC 499
>AV413010
Length = 416
Score = 55.1 bits (131), Expect = 2e-08
Identities = 35/95 (36%), Positives = 53/95 (54%), Gaps = 5/95 (5%)
Frame = +3
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTT-----RNTDFA 61
LP+EL ++IL LPV SL+RF+ VSKSW S IS+ QF K H + ++S T ++TD
Sbjct: 96 LPDELIIQILLQLPVTSLLRFKSVSKSWFSQISNPQFAKSHFNLAASPTHYLFLKSTDEF 275
Query: 62 YLQSLITSPRKSRSASTIAIPDDYDFCGTCNGLVC 96
Q+L + + + P + G+ N + C
Sbjct: 276 QFQTLDIESSPPFTTAVLNYPHEQPRSGS-NSIFC 377
>TC10709
Length = 578
Score = 54.3 bits (129), Expect = 3e-08
Identities = 35/118 (29%), Positives = 60/118 (50%), Gaps = 7/118 (5%)
Frame = +3
Query: 251 PTLGVLRDCLCICQKDKNMHFVVWQMKEFGLYKSWTLLFNIA-------GYAYDIPCRFF 303
P+L VL + + + ++H VW+M ++G +SWT L +I+ G+ Y IP
Sbjct: 12 PSLKVLGNHMYLFHDHNSIHLFVWKMVKYGALESWTQLLSISYERLHCIGFPY-IP---V 179
Query: 304 AMYMSENGDALLLSKLAVSQPQAVLYTQKDNKLEVTDVANYIFNCYAMNYIESLVPPC 361
M++ E+GD L+++ + + Y +D +E N +F A Y+ SLV PC
Sbjct: 180 PMFLLEDGDILMIA--ITENLEFIKYNIRDYSMEYIPNPNEVFWMDAYGYVPSLVSPC 347
>CB826914
Length = 575
Score = 53.5 bits (127), Expect = 5e-08
Identities = 37/127 (29%), Positives = 62/127 (48%), Gaps = 15/127 (11%)
Frame = +1
Query: 7 LPEELRLEILSWLPVKSLVRFRCVSKSWKSIISDSQFIKLHLHRSSSTTRNTDFAYLQSL 66
+ +L +EIL LPVKS+VRF+ V K W+S+ISD F LH R++ + D ++++
Sbjct: 166 MDRDLIIEILLMLPVKSIVRFKAVCKLWRSLISDPLFASLHFQRAAPSLLFADSHAIRTI 345
Query: 67 -----ITSPRKSRSASTIAIPDDYDFC---------GTCNG-LVCLHSSKYDRKDKYTSH 111
+ S R S+ + + + +D G+C G L+ S +
Sbjct: 346 DLEGPLQSHRVSQPINCHFLSNYHDLSLPRNCISIHGSCRGFLLITWISNIGYGHPWNDS 525
Query: 112 VRLWNPA 118
+ LWNP+
Sbjct: 526 LYLWNPS 546
>AV423520
Length = 468
Score = 52.4 bits (124), Expect = 1e-07
Identities = 29/50 (58%), Positives = 35/50 (70%), Gaps = 1/50 (2%)
Frame = +3
Query: 3 PPQFLPEELRLEILSWLPVKSLVRFRCVSKSWKSIIS-DSQFIKLHLHRS 51
P LP EL +EIL LPVKSL++FRCV KSW S+IS D +F + HL S
Sbjct: 153 PLPTLPFELVVEILCRLPVKSLLQFRCVCKSWNSLISGDPKFARKHLRCS 302
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.323 0.137 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,914,253
Number of Sequences: 28460
Number of extensions: 126082
Number of successful extensions: 685
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 647
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 653
length of query: 361
length of database: 4,897,600
effective HSP length: 91
effective length of query: 270
effective length of database: 2,307,740
effective search space: 623089800
effective search space used: 623089800
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0135.3


