

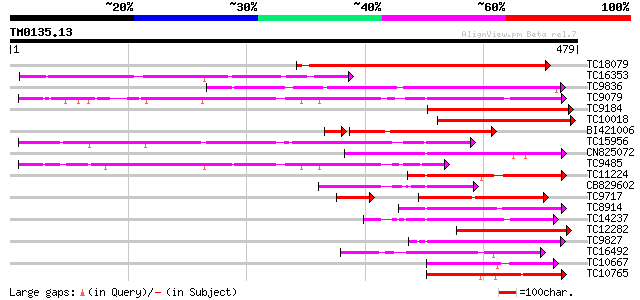
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0135.13
(479 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC18079 weakly similar to UP|UFO5_MANES (Q40287) Flavonol 3-O-gl... 202 7e-53
TC16353 weakly similar to UP|HQGT_ARATH (Q9M156) Probable hydroq... 174 4e-44
TC9836 weakly similar to UP|Q9LXV0 (Q9LXV0) Glucosyltransferase-... 150 3e-37
TC9079 weakly similar to UP|Q8W4G1 (Q8W4G1) UDP-glucose glucosyl... 147 5e-36
TC9184 weakly similar to UP|HQGT_RAUSE (Q9AR73) Hydroquinone glu... 145 1e-35
TC10018 weakly similar to UP|HQGT_RAUSE (Q9AR73) Hydroquinone gl... 135 1e-32
BI421006 114 9e-31
TC15956 weakly similar to UP|LGT_CITUN (Q9MB73) Limonoid UDP-glu... 121 3e-28
CN825072 118 2e-27
TC9485 similar to UP|Q8S999 (Q8S999) Glucosyltransferase-10, par... 115 1e-26
TC11224 weakly similar to UP|Q8S996 (Q8S996) Glucosyltransferase... 108 3e-24
CB829602 106 1e-23
TC9717 weakly similar to UP|Q9LNI1 (Q9LNI1) F6F3.22 protein, par... 77 5e-22
TC8914 similar to UP|Q9ZWS2 (Q9ZWS2) Flavonoid 3-O-galactosyl tr... 99 2e-21
TC14237 weakly similar to UP|AAR06913 (AAR06913) UDP-glycosyltra... 93 1e-19
TC12282 similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13 (Fr... 93 1e-19
TC9827 weakly similar to UP|Q8S9A2 (Q8S9A2) Glucosyltransferase-... 91 5e-19
TC16492 weakly similar to UP|AAR06919 (AAR06919) UDP-glycosyltra... 88 3e-18
TC10667 similar to UP|Q9M051 (Q9M051) Glucuronosyl transferase-l... 86 2e-17
TC10765 homologue to UP|Q9ZWQ5 (Q9ZWQ5) UDP-glycose:flavonoid gl... 85 2e-17
>TC18079 weakly similar to UP|UFO5_MANES (Q40287) Flavonol
3-O-glucosyltransferase 5 (UDP-glucose flavonoid
3-O-glucosyltransferase 5) , partial (33%)
Length = 889
Score = 202 bits (515), Expect = 7e-53
Identities = 106/217 (48%), Positives = 144/217 (65%), Gaps = 2/217 (0%)
Frame = +3
Query: 243 GPLVQEGSCETHESQGNEGETH-EYMRWLDKQEQNSVLYVSFGSAGTLTHDQFNELALGL 301
GPLV+ T ES+ GE E +RWLDKQ SV+YVSFGS GT+ Q E+ALGL
Sbjct: 3 GPLVR-----TVESKPRHGEDQDEILRWLDKQPAGSVIYVSFGSGGTMPQGQMTEIALGL 167
Query: 302 ELSGVKFLWVIRPPQKLEMIGDL-SVGHEDPLEFLPNGFLERTKGQGFVVPYWADQIEIL 360
ELS +F+WV+RPP + + G+E L +LP GF+ RT+ G VVP WA Q EIL
Sbjct: 168 ELSQHRFVWVVRPPNEADASATFFGFGNEMALNYLPEGFMNRTREFGVVVPMWAPQAEIL 347
Query: 361 GHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMIAALFSDGLKVALRPKPNEKGI 420
GH A GGF+ CGWNS+LES L+G+P+VAWPLY++QKM A + S+ L VA+R K E G+
Sbjct: 348 GHPATGGFVTPCGWNSVLESVLNGVPMVAWPLYSEQKMNAYMLSEELGVAVRVKVAEGGV 527
Query: 421 VGRGVVAEVIKNILVGEEGKEIQQRMKRLQDVAIDAL 457
V R V ++++ ++V EEG ++ R++ L+ +AL
Sbjct: 528 VCRDQVVDMVRRVMVSEEGMGMKDRVRELKLSGDEAL 638
>TC16353 weakly similar to UP|HQGT_ARATH (Q9M156) Probable hydroquinone
glucosyltransferase (Arbutin synthase) , partial (52%)
Length = 885
Score = 174 bits (440), Expect = 4e-44
Identities = 109/288 (37%), Positives = 155/288 (52%), Gaps = 6/288 (2%)
Frame = +1
Query: 9 IAIVSVPLYSHTRTILEFCKRLAHLHQDIHITCINPTVKSSCNDVKALFENLPPNIDSMF 68
+A++ P H ++EF KRL LH ++ +T I PT + ++LP +I +F
Sbjct: 73 VAMMPSPGMGHLIPMIEFAKRLI-LHHNLQVTFIIPTEAPPSKAQITVLQSLPNSISHIF 249
Query: 69 LPPVNLDDMPEKPHIPILVHATITSSLPSIYNALKTLHCSSNSIGLTSIVVDVLITQVLP 128
LPPV+L D+P+ I I + T+ SLPS+ +A + L T+++VD+ T
Sbjct: 250 LPPVDLTDLPQNAMIEIRISLTVLRSLPSLRHAFRPL-------SATALLVDLFGTDAFD 408
Query: 129 MANELNVLSYVYFSSTAMLLSLCLYPSTFDKTISC------ETVEIPGCMLIHTTDLPDP 182
+A E Y++F STA LSL LY D+ + C E V+IPG + IH +DL DP
Sbjct: 409 VAREFGASPYIFFPSTATALSLFLYLPRLDREVHCEFRELAEPVKIPGSVPIHGSDLLDP 588
Query: 183 VQNRFRCSEEYKVFLEGNKRFYLADGVIINSFFDLEPETFRALQENKGTSSTSIPHVYPV 242
VQ+ R +E YK L R+ ADG++ NSF +LEP + LQ+ + P VYPV
Sbjct: 589 VQD--RKNEAYKWVLHHATRYSQADGILENSFMELEPGAIKELQKEE----PGKPRVYPV 750
Query: 243 GPLVQEGSCETHESQGNEGETHEYMRWLDKQEQNSVLYVSFGSAGTLT 290
GPLV+ Q G E + WLD+Q SVL+V FGS GTLT
Sbjct: 751 GPLVKVDGV-----QAGSGPGSECLSWLDEQPHGSVLFVCFGSGGTLT 879
>TC9836 weakly similar to UP|Q9LXV0 (Q9LXV0) Glucosyltransferase-like
protein, partial (55%)
Length = 1286
Score = 150 bits (380), Expect = 3e-37
Identities = 98/310 (31%), Positives = 156/310 (49%), Gaps = 7/310 (2%)
Frame = +1
Query: 167 EIPGCMLIHTTDLPDPVQNRFRCSEEYKVFLEGN-KRFYLADGVIINSFFDLEPETFRAL 225
+ P IH T LP + + + +F N + +DG + N+ D + R +
Sbjct: 103 DFPEAGQIHRTQLPSNIAEADG-EDAWSLFQISNISNWVNSDGFLFNTVADFDSMGLRYV 279
Query: 226 QENKGTSSTSIPHVYPVGP-LVQEGSCETHESQGNEGETHEYMRWLDKQEQNSVLYVSFG 284
+ + +GP L+ GS + G E +WLD + NSVL+VSFG
Sbjct: 280 ARKLNRPA------WAIGPVLLSTGSGSRGKGGGISPELCR--KWLDTKPSNSVLFVSFG 435
Query: 285 SAGTLTHDQFNELALGLELSGVKFLWVIRPPQKLEMIGDLSVGHEDPLEFLPNGFLERTK 344
S T++ Q +LA L+ SG F+WV+RPP ++ + E+LP GFL R
Sbjct: 436 SMNTISASQMMQLATALDRSGRNFIWVVRPPIGFDINSEFRAE-----EWLPEGFLGRVA 600
Query: 345 GQGFVVPYWADQIEILGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMIAALFS 404
+G VV WA Q+EIL HGA+ FL HCGWNS+LES HG+PI+ WP+ A+Q +
Sbjct: 601 KKGLVVHDWAPQVEILSHGAVSAFLSHCGWNSVLESLSHGVPILGWPMAAEQFFNCKMLE 780
Query: 405 DGLKVALRPKPNEKGIVGRGVVAEVIKNIL-VGEEGKEIQQRMKRLQDVAIDALKED--- 460
+ L V + ++ V + E I+ ++ E G +I++ ++++ DA++++
Sbjct: 781 EELGVCVEVARGKRCEVRHEDLVEKIELVMNEAESGVKIRKNAGNIREMIRDAVRDEDGY 960
Query: 461 -GSSTRTITQ 469
GSS R I +
Sbjct: 961 KGSSVRAIDE 990
>TC9079 weakly similar to UP|Q8W4G1 (Q8W4G1) UDP-glucose
glucosyltransferase, partial (65%)
Length = 1585
Score = 147 bits (370), Expect = 5e-36
Identities = 134/504 (26%), Positives = 222/504 (43%), Gaps = 41/504 (8%)
Frame = +1
Query: 8 HIAIVSVPLYSHTRTILEFCKRLAHLHQDIHITCINPT------VKS-SCNDVKAL---- 56
H + PL H + + K L HL + HIT ++ +KS N + L
Sbjct: 19 HAVLTPSPLQGHINPLFQLAK-LLHL-RGFHITFVHTEYNHKRLIKSRGPNALDGLPDFV 192
Query: 57 FENLPPNID------SMFLPPVNLDDMPEKPHIPILVHATITSSLPSIYNALKTLHCSSN 110
FE +P ++ S L + D + K H+P + L LH S+
Sbjct: 193 FETIPDGLEDGDGEVSQDLYSL-CDSIRNKCHLPF-------------QDLLAKLHRSA- 327
Query: 111 SIGL----TSIVVDVLITQVLPMANELNV-LSYVYFSSTAMLLSLCLYPSTFDKTI---- 161
S GL T +V D+L+T + A +L + + + +S + +S + + DK +
Sbjct: 328 SAGLVPPVTCLVSDLLMTFTIQAAQQLQLPILLLCPASASTFMSFLHFQTLLDKGVIPLK 507
Query: 162 ----------SCETVEIPGCMLIHTTDLPDPVQNRFRCSEEYKVFLEGNKRFYLADGVII 211
+ IPG DLPD ++ + +E R A ++
Sbjct: 508 DESYLTNGYLDTKVDWIPGMQNFRLKDLPDFIRTTDPNDIMLEFMVEVADRAKRASAIVF 687
Query: 212 NSFFDLEPETFRALQENKGTSSTSIPHVYPVGPL---VQEGSCETHESQGNE--GETHEY 266
N+F +LE + AL ST +P +YP+GP + + S G+ E +
Sbjct: 688 NTFNELERDVLSAL-------STMLPSLYPIGPFPSFLNQAPQNQLASLGSNLWKEDTKC 846
Query: 267 MRWLDKQEQNSVLYVSFGSAGTLTHDQFNELALGLELSGVKFLWVIRPPQKLEMIGDLSV 326
++WL+ +E SV+YV+FGS ++ +Q E A GL FLW+IRP +++ D S
Sbjct: 847 LQWLESKEPGSVVYVNFGSITVMSPEQLLEFAWGLANGKKPFLWIIRP----DLVADGSA 1014
Query: 327 GHEDPLEFLPNGFLERTKGQGFVVPYWADQIEILGHGAIGGFLCHCGWNSMLESTLHGIP 386
L F++ T +G + W Q ++L H ++GGFL HCGWNS +ES G+P
Sbjct: 1015-------ILSPKFVDETSNRGLIAS-WCPQEKVLNHPSVGGFLTHCGWNSTIESICAGVP 1170
Query: 387 IVAWPLYADQKMIAALFSDGLKVALRPKPNEKGIVGRGVVAEVIKNILVGEEGKEIQQRM 446
++ WP ADQ + + + N K R V +I ++VG++GK+++QR
Sbjct: 1171MLCWPFVADQPTNCRYICNEWDIGIEVDTNVK----REEVENLIHELMVGDKGKKMRQRT 1338
Query: 447 KRLQDVAIDALKEDGSSTRTITQL 470
L+ A + + G S + +L
Sbjct: 1339MELKKKAEEDTRPGGCSYMNLDKL 1410
>TC9184 weakly similar to UP|HQGT_RAUSE (Q9AR73) Hydroquinone
glucosyltransferase (Arbutin synthase) , partial (26%)
Length = 643
Score = 145 bits (367), Expect = 1e-35
Identities = 67/123 (54%), Positives = 93/123 (75%)
Frame = +3
Query: 354 ADQIEILGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMIAALFSDGLKVALRP 413
A Q ++L H +IGGFL HCGWNS+LES +G+P+VAWPLYA+Q+M A L ++ +KVALRP
Sbjct: 3 APQAKVLSHASIGGFLSHCGWNSVLESVANGVPLVAWPLYAEQRMNAVLVTEDVKVALRP 182
Query: 414 KPNEKGIVGRGVVAEVIKNILVGEEGKEIQQRMKRLQDVAIDALKEDGSSTRTITQLALK 473
K E G+V +A V++ ++ GEEGK+++ +MK L++ A LKE+GSST I QLALK
Sbjct: 183 KVGENGLVELVEIARVVRTLMQGEEGKKLRCKMKDLKEAAATTLKENGSSTNQIFQLALK 362
Query: 474 WKR 476
WK+
Sbjct: 363 WKK 371
>TC10018 weakly similar to UP|HQGT_RAUSE (Q9AR73) Hydroquinone
glucosyltransferase (Arbutin synthase) , partial (24%)
Length = 586
Score = 135 bits (340), Expect = 1e-32
Identities = 59/117 (50%), Positives = 90/117 (76%)
Frame = +1
Query: 362 HGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMIAALFSDGLKVALRPKPNEKGIV 421
HG+ GGFL HCGWNS+LES ++G+P+VAWPL+A+QKM A L + +KVALRP+ E G+V
Sbjct: 1 HGSTGGFLTHCGWNSILESVVYGVPLVAWPLFAEQKMNAVLLTQEIKVALRPRVGEDGLV 180
Query: 422 GRGVVAEVIKNILVGEEGKEIQQRMKRLQDVAIDALKEDGSSTRTITQLALKWKRSA 478
+ +A V+K ++ GEEGK+++ +MK +++ +L E+GSS+ I+QLALKW +++
Sbjct: 181 EKHEIASVVKCLMEGEEGKKLRYQMKNMKEAGARSLGENGSSSNHISQLALKWSKNS 351
>BI421006
Length = 519
Score = 114 bits (285), Expect(2) = 9e-31
Identities = 57/124 (45%), Positives = 76/124 (60%)
Frame = +2
Query: 288 TLTHDQFNELALGLELSGVKFLWVIRPPQKLEMIGDLSVGHEDPLEFLPNGFLERTKGQG 347
T +Q ++A GLE S KF+WV+R K GD+ G + LP GF +R +G G
Sbjct: 116 TFIEEQIEQIANGLEQSKQKFIWVLRDADK----GDIFDGDKVKERGLPKGFEKRVEGMG 283
Query: 348 FVVPYWADQIEILGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMIAALFSDGL 407
VV WA Q+EIL H + GGF+ HCGWNS +ES G+PI AWP+++DQ L ++ L
Sbjct: 284 LVVRDWAPQLEILSHPSTGGFMSHCGWNSCIESMSMGVPIAAWPMHSDQPRNTVLITEVL 463
Query: 408 KVAL 411
KVAL
Sbjct: 464 KVAL 475
Score = 36.2 bits (82), Expect(2) = 9e-31
Identities = 15/18 (83%), Positives = 16/18 (88%)
Frame = +3
Query: 267 MRWLDKQEQNSVLYVSFG 284
M WLD+QEQ SVLYVSFG
Sbjct: 54 MEWLDRQEQKSVLYVSFG 107
>TC15956 weakly similar to UP|LGT_CITUN (Q9MB73) Limonoid
UDP-glucosyltransferase (Limonoid glucosyltransferase)
(Limonoid GTase) (LGTase) , partial (65%)
Length = 1169
Score = 121 bits (303), Expect = 3e-28
Identities = 104/399 (26%), Positives = 176/399 (44%), Gaps = 13/399 (3%)
Frame = +2
Query: 8 HIAIVSVPLYSHTRTILEFCKRLAHLHQDIHITCINPTVKSSCNDVKALFENLPPNIDS- 66
HI +VS P H +L K LA + + T S + + P I
Sbjct: 38 HILLVSFPAQGHINPLLRLAKCLAAKGSSVTLATTQ-TAGKSMRTATNITDKAPTPISDG 214
Query: 67 -----MFLPPVNLDDMPEKPHIPILVHATITSSLPSIYNALKTLHCSSNSIG------LT 115
F + DD + ++ + + I +K S+ I
Sbjct: 215 SLKFEFFDDGLGPDDDSIRGNLSLYLQQLELVGRQQILKMIKEHADSNRQISCIINNPFL 394
Query: 116 SIVVDVLITQVLPMANELNVLSYVYFSSTAMLLSLCLYPSTFDKTISCETVEIPGCMLI- 174
VVDV Q +P A S V+ + + +L +PS D V++P ++
Sbjct: 395 PWVVDVAEEQGIPSALLWIQSSAVFTAYYSYFHNLVTFPSKEDPLAD---VQLPSTSIVL 565
Query: 175 HTTDLPDPVQNRFRCSEEYKVFLEGNKRFYLADGVIINSFFDLEPETFRALQENKGTSST 234
++PD + + LE K ++++++ +LE + L S T
Sbjct: 566 KHCEIPDFLHPSCTYPFLGTLILEQFKNLNKTFCILVDTYEELEHDFIDYL------SKT 727
Query: 235 SIPHVYPVGPLVQEGSCETHESQGNEGETHEYMRWLDKQEQNSVLYVSFGSAGTLTHDQF 294
+ + PVGPL + ++ +G+ ++ + + WL+ + Q+SV+YVSFGS L +Q
Sbjct: 728 FL--IRPVGPLFKSSQAKSAAIRGDFMKSDDCIEWLNSKPQSSVVYVSFGSIVYLPQEQV 901
Query: 295 NELALGLELSGVKFLWVIRPPQKLEMIGDLSVGHEDPLEFLPNGFLERTKGQGFVVPYWA 354
+E+A GL+ S V FLWV++PP ++ + LP GFLE T +G VV W+
Sbjct: 902 DEIAFGLKKSQVSFLWVMKPPPEVTGLQP---------HVLPYGFLEETGERGRVV-QWS 1051
Query: 355 DQIEILGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLY 393
Q E+L H ++ FL HCGWNS +E+ G+P++ +P +
Sbjct: 1052PQEEVLAHPSVSCFLTHCGWNSSMEALTSGVPVLTFPAW 1168
>CN825072
Length = 712
Score = 118 bits (296), Expect = 2e-27
Identities = 71/194 (36%), Positives = 107/194 (54%), Gaps = 7/194 (3%)
Frame = -3
Query: 284 GSAGTLTHDQFNELALGLELSGVKFLWVIRPPQ-KLEMIGDLSVGHEDPLEFLPNGFLER 342
GS G+ Q E+A +E SGV+F+W +R P K M+G +D LP GFL+R
Sbjct: 710 GSRGSFDRAQITEIARAVENSGVRFVWSLRKPXLKGSMVGPSDYSVDDLASVLPEGFLDR 531
Query: 343 TKGQGFVVPYWADQIEILGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMIAAL 402
T G G V+ WA Q +L H A GGF+ HCGWNS LES G+PI WPLYA+Q+ A +
Sbjct: 530 TTGIGRVIG-WAPQTRVLTHPATGGFVSHCGWNSTLESIYFGVPIATWPLYAEQQTNAFV 354
Query: 403 FSDGLKVALRPKPNEKGIVGRG----VVAEVIKNIL--VGEEGKEIQQRMKRLQDVAIDA 456
LK+A+ + + + G + A+ I+ + V ++ E+++R+K + + +
Sbjct: 353 LVRELKIAVEISLDYRVEINGGPNYLLTADKIEGGIRSVLDKDGEVRKRVKEMSEKSRKT 174
Query: 457 LKEDGSSTRTITQL 470
L E G S + +L
Sbjct: 173 LLEGGCSYSYLDRL 132
>TC9485 similar to UP|Q8S999 (Q8S999) Glucosyltransferase-10, partial (75%)
Length = 1195
Score = 115 bits (289), Expect = 1e-26
Identities = 113/395 (28%), Positives = 177/395 (44%), Gaps = 31/395 (7%)
Frame = +1
Query: 8 HIAIVSVPLYSHTRTILEFCKRLAHLHQDIHITCINPTVKSSCNDVKALFENLPPNIDSM 67
H+ + P H +L+ K L H H+T +N + N + L P +++
Sbjct: 91 HVVCIPYPAQGHINPMLKLAK-LLHFKGGFHVTFVN----TEYNHKRLLKSRGPDSLNG- 252
Query: 68 FLPPVNLDDMPE---------KPHIPILVHATITSSLPSIYNALKTLHCSSNSIG-LTSI 117
LP + +P+ IP L +T + LP L L+ S+ + +T I
Sbjct: 253 -LPSFRFETIPDGLPETDVDVTQDIPSLCISTRKTCLPHFKKLLSKLNDVSSDVPPVTCI 429
Query: 118 VVDVLITQVLPMANELNVLSYVYFSSTAM-LLSLCLYPSTFDKTISC------------- 163
V D ++ L A ELN+ ++++++A + Y +K I
Sbjct: 430 VSDGCMSFTLDAAIELNIPEVLFWTTSACGFMGYVQYRELIEKGIIPLKDSSDITNGYLE 609
Query: 164 ETVE-IPGCMLIHTTDLPDPVQNRFRCSEEYKVFLEGN-KRFYLADGVIINSFFDLEPET 221
T+E +PG I DLP ++ +++ FL G +R A +I+N+F LE +
Sbjct: 610 TTIEWLPGMKNIRLKDLPSFLRTTDP-NDKMLDFLTGECQRALKASAIILNTFDALEHDV 786
Query: 222 FRALQENKGTSSTSIPHVYPVGPL---VQEGSCETHESQGNE--GETHEYMRWLDKQEQN 276
A S+ +P VY +GPL +++ + + S G+ E E ++WLD +E N
Sbjct: 787 LEAF-------SSILPPVYSIGPLHLLIKDVTDKNLNSLGSNLWKEDSECLKWLDTKEPN 945
Query: 277 SVLYVSFGSAGTLTHDQFNELALGLELSGVKFLWVIRPPQKLEMIGDLSVGHEDPLEFLP 336
SV+YV+FGS +T +Q E A GL S FLWVIRP DL G LP
Sbjct: 946 SVVYVNFGSITVMTSEQMVEFAWGLANSNKTFLWVIRP--------DLVAGKH---AVLP 1092
Query: 337 NGFLERTKGQGFVVPYWADQIEILGHGAIGGFLCH 371
F+ T +G + W Q ++L H AIGGFL H
Sbjct: 1093EEFVAATNDRG-RLSSWTPQEDVLTHPAIGGFLTH 1194
>TC11224 weakly similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13
(Fragment), partial (19%)
Length = 596
Score = 108 bits (269), Expect = 3e-24
Identities = 56/138 (40%), Positives = 88/138 (63%), Gaps = 4/138 (2%)
Frame = +3
Query: 337 NGFLERTKGQGFVVPYWADQIEILGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQ 396
NGFLERTK QG VV WA Q+EIL H ++G FL H GWNS+LE + G+P++ P + DQ
Sbjct: 3 NGFLERTKSQGKVVS-WAPQMEILKHASVGVFLTHGGWNSVLECIVGGVPMIGRPFFGDQ 179
Query: 397 K----MIAALFSDGLKVALRPKPNEKGIVGRGVVAEVIKNILVGEEGKEIQQRMKRLQDV 452
+ M+A L G+ + + G++ + + + +K+ + GEEGK ++Q+M L+++
Sbjct: 180 RINIWMLAKLLGVGVGL-------QNGVLAKETILKTLKSTMTGEEGKVMRQKMAELKEM 338
Query: 453 AIDALKEDGSSTRTITQL 470
A A++ DGSST+ + L
Sbjct: 339 AWKAVEPDGSSTKNLCTL 392
>CB829602
Length = 542
Score = 106 bits (264), Expect = 1e-23
Identities = 59/135 (43%), Positives = 79/135 (57%)
Frame = +1
Query: 262 ETHEYMRWLDKQEQNSVLYVSFGSAGTLTHDQFNELALGLELSGVKFLWVIRPPQKLEMI 321
E E ++WLD QE +SVLYV+FGS +T Q ELA G+ S KF+WVIRP
Sbjct: 82 EEPECLKWLDSQEPSSVLYVNFGSVINMTPQQLVELAWGIANSKKKFIWVIRP------- 240
Query: 322 GDLSVGHEDPLEFLPNGFLERTKGQGFVVPYWADQIEILGHGAIGGFLCHCGWNSMLEST 381
DL G + LP + TK +G ++ W Q ++L H A+GGFL HCGWNS +ES
Sbjct: 241 -DLVEGEASIV--LPE-IVAETKDRGIMLS-WCPQEQVLKHSALGGFLTHCGWNSTIESI 405
Query: 382 LHGIPIVAWPLYADQ 396
G+P++ P + DQ
Sbjct: 406 SSGVPLICSPFFNDQ 450
>TC9717 weakly similar to UP|Q9LNI1 (Q9LNI1) F6F3.22 protein, partial (18%)
Length = 902
Score = 77.0 bits (188), Expect(2) = 5e-22
Identities = 38/111 (34%), Positives = 67/111 (60%), Gaps = 1/111 (0%)
Frame = +2
Query: 346 QGFVVPYWADQIEILGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMIAALFSD 405
+G V WA Q+ IL H +IGGFL HCGW+S++E G P++ P + +++M+ A F +
Sbjct: 173 KGMVWRTWAPQMRILAHKSIGGFLTHCGWSSVIEGLQVGCPLIMLP-FPNEQMLVARFME 349
Query: 406 GLKVALRPKPNE-KGIVGRGVVAEVIKNILVGEEGKEIQQRMKRLQDVAID 455
G K+ ++ N+ G R VA+ ++++++ EEGK + + + L + D
Sbjct: 350 GKKLGVKVSKNQHDGKFTRDSVAKALRSVMLEEEGKSYRCQAEELSKIVGD 502
Score = 44.3 bits (103), Expect(2) = 5e-22
Identities = 21/32 (65%), Positives = 26/32 (80%)
Frame = +1
Query: 277 SVLYVSFGSAGTLTHDQFNELALGLELSGVKF 308
SV+YV+FGS TL+++ F ELALGLELSG F
Sbjct: 4 SVIYVAFGSEVTLSNEDFTELALGLELSGFPF 99
>TC8914 similar to UP|Q9ZWS2 (Q9ZWS2) Flavonoid 3-O-galactosyl transferase,
partial (32%)
Length = 627
Score = 98.6 bits (244), Expect = 2e-21
Identities = 49/142 (34%), Positives = 83/142 (57%)
Frame = +2
Query: 329 EDPLEFLPNGFLERTKGQGFVVPYWADQIEILGHGAIGGFLCHCGWNSMLESTLHGIPIV 388
+ P LPNGF+ERTK G +VP WA Q +ILGH ++G F+ HCG NS+ ES +G+P++
Sbjct: 11 DHPKSLLPNGFIERTKNSGKIVP-WAPQTQILGHESVGVFVTHCGCNSVFESISNGVPMI 187
Query: 389 AWPLYADQKMIAALFSDGLKVALRPKPNEKGIVGRGVVAEVIKNILVGEEGKEIQQRMKR 448
P + D M + D ++ ++ E G+ + + + + ILV EEGK ++++ ++
Sbjct: 188 CRPFFGDHGMAGRIIEDIWEIGVK---IEGGVFTQDGLVKSLNLILVQEEGKIMREKAQK 358
Query: 449 LQDVAIDALKEDGSSTRTITQL 470
++ +DA G +T+ L
Sbjct: 359 VKKTVLDAAGPQGKATQDFKTL 424
>TC14237 weakly similar to UP|AAR06913 (AAR06913) UDP-glycosyltransferase
85A8, partial (27%)
Length = 999
Score = 92.8 bits (229), Expect = 1e-19
Identities = 52/164 (31%), Positives = 89/164 (53%)
Frame = +2
Query: 300 GLELSGVKFLWVIRPPQKLEMIGDLSVGHEDPLEFLPNGFLERTKGQGFVVPYWADQIEI 359
G+ S V FLW++RP D+ +G ED + +P FL+ KG+G++ W Q E+
Sbjct: 194 GIANSKVPFLWILRP--------DVVMG-EDSIN-VPQEFLDEIKGRGYIAS-WCFQEEV 340
Query: 360 LGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMIAALFSDGLKVALRPKPNEKG 419
L H +IG FL HCGWNS +E G+P++ WP +++Q + ++ + + K
Sbjct: 341 LSHPSIGAFLTHCGWNSTIEGISAGLPLICWPFFSEQHTNSRYACTTWEIGMEVNHDVK- 517
Query: 420 IVGRGVVAEVIKNILVGEEGKEIQQRMKRLQDVAIDALKEDGSS 463
R + ++ ++ GE+GKE++++ + AI+A GSS
Sbjct: 518 ---RDEITTLVNEMMKGEKGKEMRKKGLEWKKKAIEATGLGGSS 640
>TC12282 similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13 (Fragment),
partial (7%)
Length = 511
Score = 92.8 bits (229), Expect = 1e-19
Identities = 46/98 (46%), Positives = 67/98 (67%), Gaps = 1/98 (1%)
Frame = +3
Query: 378 LESTLHGIPIVAWPLYADQKMIAALFSDGLKVALRPKPN-EKGIVGRGVVAEVIKNILVG 436
LES ++G+P++ WPL+A+Q+M A + +D LK+ +RPK + E G+V R +A +K I+ G
Sbjct: 6 LESVVNGVPMIVWPLFAEQRMNAKIITDALKIGVRPKVDGETGMVKREEIARGVKRIMEG 185
Query: 437 EEGKEIQQRMKRLQDVAIDALKEDGSSTRTITQLALKW 474
+E EI +R+K L D A AL E GSS I+ LALKW
Sbjct: 186 DESLEICRRVKELSDGAAAALSEHGSSRNAISSLALKW 299
>TC9827 weakly similar to UP|Q8S9A2 (Q8S9A2) Glucosyltransferase-7
(Fragment), partial (42%)
Length = 611
Score = 90.5 bits (223), Expect = 5e-19
Identities = 51/132 (38%), Positives = 74/132 (55%)
Frame = +2
Query: 338 GFLERTKGQGFVVPYWADQIEILGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQK 397
GF+E T+ +G +V W Q+ +L H A+G FL HCGWNS LES G+P++A PL+ D
Sbjct: 2 GFVETTE-KGLIVT-WCPQLLVLKHQALGCFLTHCGWNSTLESVSLGVPVIAMPLWTDPV 175
Query: 398 MIAALFSDGLKVALRPKPNEKGIVGRGVVAEVIKNILVGEEGKEIQQRMKRLQDVAIDAL 457
L D K +R +EK IV R + IK I+ E+ EI+ + ++A D+L
Sbjct: 176 TNGKLIVDVWKTGVRAVADEKEIVRRETIKNCIKEIIETEKINEIKSNAIKWMNLAKDSL 355
Query: 458 KEDGSSTRTITQ 469
E G S + I +
Sbjct: 356 AEGGRSDKNIAE 391
>TC16492 weakly similar to UP|AAR06919 (AAR06919) UDP-glycosyltransferase
88B1, partial (8%)
Length = 750
Score = 88.2 bits (217), Expect = 3e-18
Identities = 62/177 (35%), Positives = 86/177 (48%), Gaps = 4/177 (2%)
Frame = +1
Query: 280 YVSFGSAGTLTHDQFNELALGLELSGVKFLWVIRPPQKLEMIGDLSVGHEDPLEFLPNGF 339
++ FGS L+ ELA G+ELSG+ F W + K G L LP GF
Sbjct: 1 FIGFGSELKLSQQDLTELAHGIELSGLPFFWAL----KYLKDGSLE---------LPEGF 141
Query: 340 LERTKGQGFVVPYWADQIEILGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMI 399
+RTK +G V WA Q +IL HG IGG + H S++E G +V P Y DQ
Sbjct: 142 EDRTKDRGIVWKTWAPQPKILAHGVIGGCMSHSAAGSVIEMVNFGHVLVTLPYYLDQ--- 312
Query: 400 AALFSDGL---KVALR-PKPNEKGIVGRGVVAEVIKNILVGEEGKEIQQRMKRLQDV 452
LFS L KV + P+ + G R VA+ ++ +V EEG ++ K + +V
Sbjct: 313 -CLFSRALEEKKVGIEVPRNEQDGSFTRESVAKTLRLAIVDEEGSVYRKNAKEMGNV 480
>TC10667 similar to UP|Q9M051 (Q9M051) Glucuronosyl transferase-like
protein, partial (20%)
Length = 824
Score = 85.5 bits (210), Expect = 2e-17
Identities = 47/113 (41%), Positives = 66/113 (57%), Gaps = 2/113 (1%)
Frame = +1
Query: 353 WADQIEILGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQKMIAALFSDGLKVA-- 410
WA Q ++L H AIG F HCGWNS LES G+P++ P + DQK+ A SD KV
Sbjct: 22 WAPQEQVLKHPAIGAFWTHCGWNSTLESVCEGVPMICSPCFGDQKVNAKYVSDVWKVGVQ 201
Query: 411 LRPKPNEKGIVGRGVVAEVIKNILVGEEGKEIQQRMKRLQDVAIDALKEDGSS 463
L+ KP GRG + + I +++G+E EI+ + +L++ A L E GSS
Sbjct: 202 LQNKP------GRGEIEKTISLLMLGDEAYEIRGNILKLKEKANVCLSEGGSS 342
>TC10765 homologue to UP|Q9ZWQ5 (Q9ZWQ5) UDP-glycose:flavonoid
glycosyltransferase, partial (19%)
Length = 542
Score = 85.1 bits (209), Expect = 2e-17
Identities = 47/127 (37%), Positives = 77/127 (60%), Gaps = 9/127 (7%)
Frame = +2
Query: 353 WADQIEILGHGAIGGFLCHCGWNSMLESTLHGIPIVAWPLYADQ----KMIAALFSDGLK 408
WA Q IL H A GGFL HCGWN+++E+ G+P++ P ++DQ K+I + G++
Sbjct: 8 WAPQPLILNHLATGGFLTHCGWNAVVEAISAGVPMITMPGFSDQYYNEKLITEVHGFGVE 187
Query: 409 V-----ALRPKPNEKGIVGRGVVAEVIKNILVGEEGKEIQQRMKRLQDVAIDALKEDGSS 463
V ++ P +K +V + + +K+ L+GEEG EI+ + K +QD A A+++ GSS
Sbjct: 188 VGAAEWSISPYDGKKKVVSGERIEKAVKS-LMGEEGAEIRSKAKEVQDKAWKAVQQGGSS 364
Query: 464 TRTITQL 470
++T L
Sbjct: 365 YNSLTVL 385
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.137 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,867,850
Number of Sequences: 28460
Number of extensions: 132599
Number of successful extensions: 818
Number of sequences better than 10.0: 94
Number of HSP's better than 10.0 without gapping: 780
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 785
length of query: 479
length of database: 4,897,600
effective HSP length: 94
effective length of query: 385
effective length of database: 2,222,360
effective search space: 855608600
effective search space used: 855608600
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0135.13


