

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0134.5
(510 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
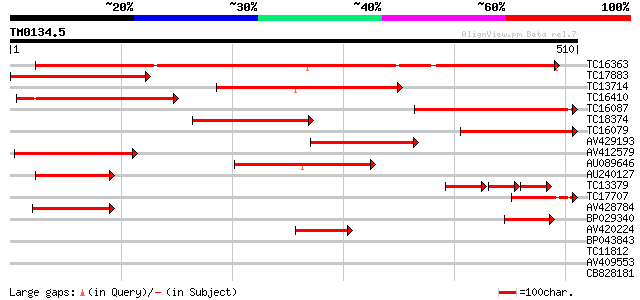
Score E
Sequences producing significant alignments: (bits) Value
TC16363 UP|Q84RQ3 (Q84RQ3) Sucrose transporter 4 protein, complete 502 e-143
TC17883 similar to UP|Q41152 (Q41152) Sucrose carrier, partial (... 256 8e-69
TC13714 homologue to UP|Q7XA53 (Q7XA53) Sucrose transporter, par... 229 1e-60
TC16410 similar to UP|Q9XHL6 (Q9XHL6) Sucrose transport protein ... 224 3e-59
TC16087 similar to UP|Q40583 (Q40583) Sucrose transporter, parti... 223 4e-59
TC18374 similar to UP|Q9FIX9 (Q9FIX9) Sucrose transporter protei... 222 1e-58
TC16079 similar to UP|O04077 (O04077) Sucrose transport protein,... 205 1e-53
AV429193 194 2e-50
AV412579 160 5e-40
AU089646 129 9e-31
AU240127 112 1e-25
TC13379 similar to UP|O65803 (O65803) Sucrose/H+ symporter, part... 54 1e-22
TC17707 similar to UP|Q7XA53 (Q7XA53) Sucrose transporter, parti... 76 1e-14
AV428784 72 3e-13
BP029340 61 5e-10
AV420224 59 2e-09
BP043843 30 0.72
TC11812 similar to UP|Q93W01 (Q93W01) At2g01080/F23H14.5, partia... 30 1.2
AV409553 29 2.1
CB828181 29 2.1
>TC16363 UP|Q84RQ3 (Q84RQ3) Sucrose transporter 4 protein, complete
Length = 2164
Score = 502 bits (1293), Expect = e-143
Identities = 253/483 (52%), Positives = 325/483 (66%), Gaps = 12/483 (2%)
Frame = +3
Query: 24 PQQSSPLRKMIAVASIAAGIQFGWALQLSLLTPYVQTLGVPHIWASFIWLCGPVSGLLVQ 83
P PLR+++ VAS+A+GIQFGWALQLSLLTPYVQ LG+PH WAS IWLCGPVSGL VQ
Sbjct: 246 PPARVPLRQLLRVASVASGIQFGWALQLSLLTPYVQQLGIPHQWASIIWLCGPVSGLFVQ 425
Query: 84 PIVGYYSDRTTCRFGRRRPFILAGAVAVAISVFLIGYAADIGHSMGDDLAKKTRPRAVAI 143
P+VG+ SD+ T RFGRRRPFILAGA ++ ++V +IGYAADIG +GD + RP A+ +
Sbjct: 426 PLVGHLSDKCTSRFGRRRPFILAGAASIVVAVLIIGYAADIGWMLGD--TESFRPAAITV 599
Query: 144 FVFGFWILDVANNMLQGPCRAFLGDLAAGDQKKTRTAMGFFSFFMAVGNVLGYAAGAYSG 203
FV GFWILDVANN+ QGPCRA L DL + D ++TR A +FS FMA+GN+LGYA GAYSG
Sbjct: 600 FVIGFWILDVANNVTQGPCRALLADLTSKDNRRTRVANAYFSLFMAIGNILGYATGAYSG 779
Query: 204 LHKIFPFTVTDACPSFCANLKSCFFFSILLLLILSIAALIYVEDTPLTKKPAADVDSPVS 263
++IF FT++ AC CANLKS FF + + + + ++ + PL AA
Sbjct: 780 WYRIFTFTLSPACTISCANLKSAFFLDVAFIAVTTYVSITAAHEVPLNSSGAAHAGEGAG 959
Query: 264 CFG--------DLFGAFKELKKPMWILMLVTAVNWVAWFPFFLFDTDWMGREVYGG-VVG 314
G +LFG FK +WI++ VTA+NW WFPF LFDTDWMGRE+YG G
Sbjct: 960 ESGSTEEAFMWELFGTFKYFSSTVWIILSVTALNWTGWFPFILFDTDWMGREIYGADPNG 1139
Query: 315 EKAYDDGVRAGSLGLMINAVVLAFMSLAVEPLGRILGGVKNLWGIVNFILAICMAMTVLI 374
YD GVR G+LGLM+N+VVL SL +E L R G +WGI N ++A+C +++
Sbjct: 1140GPNYDAGVRMGALGLMLNSVVLGVTSLLMEKLCR-KRGAGFVWGISNILMAVCFLAMLVV 1316
Query: 375 TKTAEHDRLISGGATVGAPSSGVKAGAIAFFGVMGIPLAINFSVPFALASIYSSAAGAGQ 434
T A G P +G+ A+ F ++G PLAI +SVP+AL S ++ G GQ
Sbjct: 1317TYVAN----TIGYVGKDLPPTGIVIAALIIFTILGFPLAITYSVPYALISKHTEPLGLGQ 1484
Query: 435 GLSLGVLNLAIVVPQMLVSTLSGPWDALFGGGNLPAFVVGAVMAAVSAIMAIVLLP---T 491
GLS+GVLNLAIV+PQ++VS SGPWD LFGGGN AF VGAV A +S ++A++ +P T
Sbjct: 1485GLSMGVLNLAIVIPQIVVSLGSGPWDQLFGGGNSAAFAVGAVAAIMSGLLAVLAIPRTGT 1664
Query: 492 PKP 494
KP
Sbjct: 1665QKP 1673
>TC17883 similar to UP|Q41152 (Q41152) Sucrose carrier, partial (19%)
Length = 428
Score = 256 bits (653), Expect = 8e-69
Identities = 126/126 (100%), Positives = 126/126 (100%)
Frame = +3
Query: 1 MESPTKSNHLDTKGSSLQIEAHIPQQSSPLRKMIAVASIAAGIQFGWALQLSLLTPYVQT 60
MESPTKSNHLDTKGSSLQIEAHIPQQSSPLRKMIAVASIAAGIQFGWALQLSLLTPYVQT
Sbjct: 51 MESPTKSNHLDTKGSSLQIEAHIPQQSSPLRKMIAVASIAAGIQFGWALQLSLLTPYVQT 230
Query: 61 LGVPHIWASFIWLCGPVSGLLVQPIVGYYSDRTTCRFGRRRPFILAGAVAVAISVFLIGY 120
LGVPHIWASFIWLCGPVSGLLVQPIVGYYSDRTTCRFGRRRPFILAGAVAVAISVFLIGY
Sbjct: 231 LGVPHIWASFIWLCGPVSGLLVQPIVGYYSDRTTCRFGRRRPFILAGAVAVAISVFLIGY 410
Query: 121 AADIGH 126
AADIGH
Sbjct: 411 AADIGH 428
>TC13714 homologue to UP|Q7XA53 (Q7XA53) Sucrose transporter, partial (32%)
Length = 512
Score = 229 bits (583), Expect = 1e-60
Identities = 110/170 (64%), Positives = 134/170 (78%), Gaps = 3/170 (1%)
Frame = +2
Query: 187 FMAVGNVLGYAAGAYSGLHKIFPFTVTDACPSFCANLKSCFFFSILLLLILSIAALIYVE 246
FMAVGNVLGYAAGAYS L+ +FPFT T AC +CANLKSCFF SI LL L+ AAL+YV+
Sbjct: 2 FMAVGNVLGYAAGAYSKLYHVFPFTKTTACDVYCANLKSCFFLSIALLTTLATAALVYVK 181
Query: 247 DTPLTKKPA---ADVDSPVSCFGDLFGAFKELKKPMWILMLVTAVNWVAWFPFFLFDTDW 303
+ PL+ + A +D + + CFG L GAF+ELK+PMWIL+LVT +NW+AWFPF LFDTDW
Sbjct: 182 EVPLSPEKAVIDSDDNGGMPCFGQLSGAFRELKRPMWILLLVTCLNWIAWFPFLLFDTDW 361
Query: 304 MGREVYGGVVGEKAYDDGVRAGSLGLMINAVVLAFMSLAVEPLGRILGGV 353
MG+EVYGG VG+ AYD GV AG+LGLM+N+VVL SL +E L R +GGV
Sbjct: 362 MGKEVYGGEVGDHAYDQGVHAGALGLMLNSVVLGATSLGLEYLARGVGGV 511
>TC16410 similar to UP|Q9XHL6 (Q9XHL6) Sucrose transport protein SUT1,
partial (27%)
Length = 502
Score = 224 bits (571), Expect = 3e-59
Identities = 108/146 (73%), Positives = 126/146 (85%)
Frame = +2
Query: 7 SNHLDTKGSSLQIEAHIPQQSSPLRKMIAVASIAAGIQFGWALQLSLLTPYVQTLGVPHI 66
S H + K SSL +EA P ++SPLRK+I VASIAAG+QFGWALQLSLLTPYVQ LG+PH
Sbjct: 68 SKHNNLKPSSLHVEAP-PPEASPLRKIIVVASIAAGVQFGWALQLSLLTPYVQLLGIPHK 244
Query: 67 WASFIWLCGPVSGLLVQPIVGYYSDRTTCRFGRRRPFILAGAVAVAISVFLIGYAADIGH 126
WA++IWLCGP+SG+LVQPIVGY+SDR T RFGRRRPFI AGA+AVAI+VFLIGYAAD GH
Sbjct: 245 WAAYIWLCGPISGMLVQPIVGYHSDRCTSRFGRRRPFIAAGALAVAIAVFLIGYAADFGH 424
Query: 127 SMGDDLAKKTRPRAVAIFVFGFWILD 152
++GD L KK R RA+AIF+ GFWILD
Sbjct: 425 ALGDRLDKKIRTRAIAIFIVGFWILD 502
>TC16087 similar to UP|Q40583 (Q40583) Sucrose transporter, partial (23%)
Length = 623
Score = 223 bits (569), Expect = 4e-59
Identities = 109/146 (74%), Positives = 130/146 (88%)
Frame = +2
Query: 365 AICMAMTVLITKTAEHDRLISGGATVGAPSSGVKAGAIAFFGVMGIPLAINFSVPFALAS 424
A+CMAMT+LITK AEHDR +SGGAT+G P+ GVKAGA+ FF V+GIPLAI +SVPFALAS
Sbjct: 2 AVCMAMTMLITKVAEHDRRVSGGATIGRPTPGVKAGALMFFAVLGIPLAITYSVPFALAS 181
Query: 425 IYSSAAGAGQGLSLGVLNLAIVVPQMLVSTLSGPWDALFGGGNLPAFVVGAVMAAVSAIM 484
IYSS+ GAGQGLSLGVLN+AIV+PQM+VS L+GPWD+LFGGGNLPAF+VGAVMAAVSA++
Sbjct: 182 IYSSSTGAGQGLSLGVLNVAIVIPQMIVSALNGPWDSLFGGGNLPAFMVGAVMAAVSAVL 361
Query: 485 AIVLLPTPKPADVAKASSLPPGGGFH 510
A+VLLP+PKP ++AKAS GFH
Sbjct: 362 AMVLLPSPKPEEMAKASI--SASGFH 433
>TC18374 similar to UP|Q9FIX9 (Q9FIX9) Sucrose transporter protein, partial
(17%)
Length = 330
Score = 222 bits (566), Expect = 1e-58
Identities = 109/109 (100%), Positives = 109/109 (100%)
Frame = +3
Query: 165 FLGDLAAGDQKKTRTAMGFFSFFMAVGNVLGYAAGAYSGLHKIFPFTVTDACPSFCANLK 224
FLGDLAAGDQKKTRTAMGFFSFFMAVGNVLGYAAGAYSGLHKIFPFTVTDACPSFCANLK
Sbjct: 3 FLGDLAAGDQKKTRTAMGFFSFFMAVGNVLGYAAGAYSGLHKIFPFTVTDACPSFCANLK 182
Query: 225 SCFFFSILLLLILSIAALIYVEDTPLTKKPAADVDSPVSCFGDLFGAFK 273
SCFFFSILLLLILSIAALIYVEDTPLTKKPAADVDSPVSCFGDLFGAFK
Sbjct: 183 SCFFFSILLLLILSIAALIYVEDTPLTKKPAADVDSPVSCFGDLFGAFK 329
>TC16079 similar to UP|O04077 (O04077) Sucrose transport protein, partial
(18%)
Length = 573
Score = 205 bits (522), Expect = 1e-53
Identities = 105/105 (100%), Positives = 105/105 (100%)
Frame = +2
Query: 406 GVMGIPLAINFSVPFALASIYSSAAGAGQGLSLGVLNLAIVVPQMLVSTLSGPWDALFGG 465
GVMGIPLAINFSVPFALASIYSSAAGAGQGLSLGVLNLAIVVPQMLVSTLSGPWDALFGG
Sbjct: 2 GVMGIPLAINFSVPFALASIYSSAAGAGQGLSLGVLNLAIVVPQMLVSTLSGPWDALFGG 181
Query: 466 GNLPAFVVGAVMAAVSAIMAIVLLPTPKPADVAKASSLPPGGGFH 510
GNLPAFVVGAVMAAVSAIMAIVLLPTPKPADVAKASSLPPGGGFH
Sbjct: 182 GNLPAFVVGAVMAAVSAIMAIVLLPTPKPADVAKASSLPPGGGFH 316
>AV429193
Length = 295
Score = 194 bits (494), Expect = 2e-50
Identities = 94/97 (96%), Positives = 94/97 (96%)
Frame = +1
Query: 271 AFKELKKPMWILMLVTAVNWVAWFPFFLFDTDWMGREVYGGVVGEKAYDDGVRAGSLGLM 330
AFKELKKPMWILMLVTAVNWVAWFPFFLFDTDWMGREVYGGVVGEKAYDDGVRAGSLGLM
Sbjct: 4 AFKELKKPMWILMLVTAVNWVAWFPFFLFDTDWMGREVYGGVVGEKAYDDGVRAGSLGLM 183
Query: 331 INAVVLAFMSLAVEPLGRILGGVKNLWGIVNFILAIC 367
INAVVLAFMSLA EPL RILGGVKNLW IVNFILAIC
Sbjct: 184 INAVVLAFMSLAAEPLWRILGGVKNLWRIVNFILAIC 294
>AV412579
Length = 394
Score = 160 bits (405), Expect = 5e-40
Identities = 82/112 (73%), Positives = 90/112 (80%), Gaps = 1/112 (0%)
Frame = +3
Query: 5 TKSNHLDTKGSSLQIE-AHIPQQSSPLRKMIAVASIAAGIQFGWALQLSLLTPYVQTLGV 63
T N+ SS Q+E A SPL KMIAVASIAAG+QFGWALQLSLLTPYVQ LGV
Sbjct: 57 TTQNNNSIAPSSFQVEPAQHAAGPSPLAKMIAVASIAAGVQFGWALQLSLLTPYVQLLGV 236
Query: 64 PHIWASFIWLCGPVSGLLVQPIVGYYSDRTTCRFGRRRPFILAGAVAVAISV 115
PH W+SFIWLCGP+SGL+VQPIVGY SDR T RFGRRRPFI AGA+AVAI+V
Sbjct: 237 PHKWSSFIWLCGPISGLVVQPIVGYSSDRCTSRFGRRRPFIFAGAIAVAIAV 392
>AU089646
Length = 411
Score = 129 bits (325), Expect = 9e-31
Identities = 62/136 (45%), Positives = 83/136 (60%), Gaps = 9/136 (6%)
Frame = +2
Query: 203 GLHKIFPFTVTDACPSFCANLKSCFFFSILLLLILSIAALIYVEDTPLTKKPAADVDSPV 262
G +K+FPFT+T AC CANLKS FF I+ ++I + + + + PL A V+
Sbjct: 2 GWYKVFPFTLTPACNVSCANLKSAFFLDIIFIVITTYISTVSAHEVPLNSSGAHPVEEGA 181
Query: 263 --------SCFGDLFGAFKELKKPMWILMLVTAVNWVAWFPFFLFDTDWMGREVYGGVVG 314
+ ++FG FK KP+WI++ VTA+ W+ WFPF LFDTDWMGRE+YGG
Sbjct: 182 GESXSAEEAFLWEMFGTFKYFSKPIWIILSVTALTWIGWFPFLLFDTDWMGREIYGGEPN 361
Query: 315 E-KAYDDGVRAGSLGL 329
E YD GVR G+LGL
Sbjct: 362 EGPNYDIGVRMGALGL 409
>AU240127
Length = 300
Score = 112 bits (281), Expect = 1e-25
Identities = 50/71 (70%), Positives = 59/71 (82%)
Frame = +3
Query: 24 PQQSSPLRKMIAVASIAAGIQFGWALQLSLLTPYVQTLGVPHIWASFIWLCGPVSGLLVQ 83
P+ PL +++ VAS+A GIQFGWALQLSLLTPYVQ LG+PH WAS IWLCGP+SGL VQ
Sbjct: 75 PRARVPLTQLLKVASVAGGIQFGWALQLSLLTPYVQQLGIPHAWASIIWLCGPLSGLFVQ 254
Query: 84 PIVGYYSDRTT 94
P+VG+ SDR T
Sbjct: 255 PLVGHMSDRCT 287
>TC13379 similar to UP|O65803 (O65803) Sucrose/H+ symporter, partial (12%)
Length = 534
Score = 53.9 bits (128), Expect(3) = 1e-22
Identities = 24/37 (64%), Positives = 31/37 (82%)
Frame = +2
Query: 393 PSSGVKAGAIAFFGVMGIPLAINFSVPFALASIYSSA 429
PS+GVKA A+ F V+GIPLAI +S+PFAL SIYS++
Sbjct: 2 PSTGVKASAMILFSVLGIPLAITYSIPFALGSIYSTS 112
Score = 51.6 bits (122), Expect(3) = 1e-22
Identities = 24/28 (85%), Positives = 27/28 (95%)
Frame = +1
Query: 460 DALFGGGNLPAFVVGAVMAAVSAIMAIV 487
DALFGGGNLPAF+VGA+MA VSAI+AIV
Sbjct: 205 DALFGGGNLPAFLVGAIMAVVSAILAIV 288
Score = 37.7 bits (86), Expect(3) = 1e-22
Identities = 16/28 (57%), Positives = 22/28 (78%)
Frame = +3
Query: 431 GAGQGLSLGVLNLAIVVPQMLVSTLSGP 458
G GQGL GV N+A+V+PQ+++ LSGP
Sbjct: 117 GGGQGLYFGVHNIAMVIPQIIIWGLSGP 200
>TC17707 similar to UP|Q7XA53 (Q7XA53) Sucrose transporter, partial (11%)
Length = 554
Score = 75.9 bits (185), Expect = 1e-14
Identities = 39/59 (66%), Positives = 47/59 (79%)
Frame = +2
Query: 452 VSTLSGPWDALFGGGNLPAFVVGAVMAAVSAIMAIVLLPTPKPADVAKASSLPPGGGFH 510
VS SG WD LFGGGNLPAFVVGAV AA+S I+++VLLP+P P D+ KA++ GGGFH
Sbjct: 2 VSVSSGNWDDLFGGGNLPAFVVGAVAAAISGILSMVLLPSP-PPDLVKAAT-ASGGGFH 172
>AV428784
Length = 334
Score = 71.6 bits (174), Expect = 3e-13
Identities = 39/74 (52%), Positives = 47/74 (62%)
Frame = +2
Query: 21 AHIPQQSSPLRKMIAVASIAAGIQFGWALQLSLLTPYVQTLGVPHIWASFIWLCGPVSGL 80
A I Q + K++ ASIA +QFGWALQ SLLTP VQ LGVP+ S I GP+ GL
Sbjct: 113 AQIAQGPTQFAKIMVGASIAPEVQFGWALQFSLLTPNVQLLGVPNKGPSLIGFWGPIFGL 292
Query: 81 LVQPIVGYYSDRTT 94
+VQP+ Y DR T
Sbjct: 293 VVQPMGAYSRDR*T 334
>BP029340
Length = 402
Score = 60.8 bits (146), Expect = 5e-10
Identities = 27/45 (60%), Positives = 34/45 (75%)
Frame = -2
Query: 446 VVPQMLVSTLSGPWDALFGGGNLPAFVVGAVMAAVSAIMAIVLLP 490
V PQM+VS SGPWD LFGGGN PAF V AV A +S ++A++ +P
Sbjct: 401 VFPQMVVSLGSGPWDQLFGGGNSPAFAVAAVAALISGLIAVLAIP 267
>AV420224
Length = 241
Score = 58.9 bits (141), Expect = 2e-09
Identities = 24/51 (47%), Positives = 35/51 (68%)
Frame = +1
Query: 258 VDSPVSCFGDLFGAFKELKKPMWILMLVTAVNWVAWFPFFLFDTDWMGREV 308
+D P + +L + + L M +++V A+ W++WFPFFLFDTDWMGREV
Sbjct: 88 MDGPGAVLVNLLTSLRHLPPAMHSVLIVMALTWLSWFPFFLFDTDWMGREV 240
>BP043843
Length = 477
Score = 30.4 bits (67), Expect = 0.72
Identities = 16/28 (57%), Positives = 22/28 (78%)
Frame = -1
Query: 483 IMAIVLLPTPKPADVAKASSLPPGGGFH 510
I+++VLLP+P P D+AKA + GGGFH
Sbjct: 477 ILSLVLLPSP-PPDLAKAVT-AAGGGFH 400
>TC11812 similar to UP|Q93W01 (Q93W01) At2g01080/F23H14.5, partial (52%)
Length = 592
Score = 29.6 bits (65), Expect = 1.2
Identities = 20/69 (28%), Positives = 35/69 (49%), Gaps = 5/69 (7%)
Frame = +1
Query: 197 AAGAYSGLHKIFP--FTVTDACPSFCANLKS---CFFFSILLLLILSIAALIYVEDTPLT 251
A+ + +G H+ P + T + PS A+LK C F + L +L++A ++ +
Sbjct: 139 ASASSNGYHQPHPPYYPRTPSRPSSSASLKGCCCCLFLLLSFLALLALAVVLVIILAVKP 318
Query: 252 KKPAADVDS 260
KKP D+ S
Sbjct: 319 KKPQFDLQS 345
>AV409553
Length = 433
Score = 28.9 bits (63), Expect = 2.1
Identities = 16/47 (34%), Positives = 24/47 (51%), Gaps = 3/47 (6%)
Frame = +2
Query: 45 FGWALQLSLLTPYVQTLGVPHIWASFIWLCG---PVSGLLVQPIVGY 88
F W + S L + TLG+ +I + W G P SGLL+ ++ Y
Sbjct: 257 FNWWMFTSFLGALIATLGLVYIQENLGWGLGYGIPTSGLLLSLVIFY 397
>CB828181
Length = 562
Score = 28.9 bits (63), Expect = 2.1
Identities = 11/24 (45%), Positives = 19/24 (78%)
Frame = +2
Query: 222 NLKSCFFFSILLLLILSIAALIYV 245
N+ +C FFS+LL+L+L + + +YV
Sbjct: 377 NVVTCCFFSLLLVLVLDLCS*LYV 448
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.326 0.141 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,058,386
Number of Sequences: 28460
Number of extensions: 132593
Number of successful extensions: 1004
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 988
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 995
length of query: 510
length of database: 4,897,600
effective HSP length: 94
effective length of query: 416
effective length of database: 2,222,360
effective search space: 924501760
effective search space used: 924501760
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0134.5


