

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
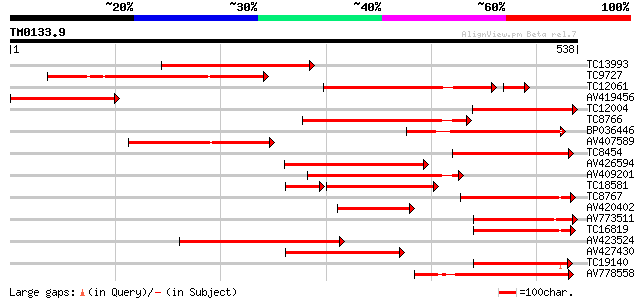
Query= TM0133.9
(538 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC13993 similar to UP|Q8VWP9 (Q8VWP9) Fiddlehead-like protein, p... 285 1e-77
TC9727 weakly similar to UP|Q9FV67 (Q9FV67) Fatty acid elongase ... 227 3e-60
TC12061 similar to UP|Q9LN49 (Q9LN49) F18O14.21, partial (35%) 204 2e-57
AV419456 207 3e-54
TC12004 similar to UP|Q8VWP9 (Q8VWP9) Fiddlehead-like protein, p... 202 1e-52
TC8766 similar to UP|Q8LAZ1 (Q8LAZ1) Very-long-chain fatty acid ... 197 5e-51
BP036446 184 4e-47
AV407589 181 2e-46
TC8454 similar to UP|Q84UX3 (Q84UX3) Fatty acid elongase (Fragme... 178 2e-45
AV426594 176 7e-45
AV409201 174 2e-44
TC18581 similar to UP|Q9FG87 (Q9FG87) Beta-ketoacyl-CoA synthase... 144 1e-43
TC8767 homologue to UP|Q8LAZ1 (Q8LAZ1) Very-long-chain fatty aci... 158 2e-39
AV420402 150 4e-37
AV773511 138 2e-33
TC16819 similar to UP|Q8LAZ1 (Q8LAZ1) Very-long-chain fatty acid... 138 3e-33
AV423524 137 6e-33
AV427430 135 2e-32
TC19140 similar to PIR|F86141|F86141 protein T25K16.11 [imported... 131 3e-31
AV778558 130 7e-31
>TC13993 similar to UP|Q8VWP9 (Q8VWP9) Fiddlehead-like protein, partial
(26%)
Length = 437
Score = 285 bits (729), Expect = 1e-77
Identities = 145/145 (100%), Positives = 145/145 (100%)
Frame = +3
Query: 145 YIELASKSGKFDEESLAFQKRILMSSGIGDETYIPKAVVSSTTNTATMKEGRAEASTVMF 204
YIELASKSGKFDEESLAFQKRILMSSGIGDETYIPKAVVSSTTNTATMKEGRAEASTVMF
Sbjct: 3 YIELASKSGKFDEESLAFQKRILMSSGIGDETYIPKAVVSSTTNTATMKEGRAEASTVMF 182
Query: 205 GALDELFEKTGIRPKDIGILVVNCSIFNPTPSLSAMIINHYKMRGNILSYNLGGMGCSAG 264
GALDELFEKTGIRPKDIGILVVNCSIFNPTPSLSAMIINHYKMRGNILSYNLGGMGCSAG
Sbjct: 183 GALDELFEKTGIRPKDIGILVVNCSIFNPTPSLSAMIINHYKMRGNILSYNLGGMGCSAG 362
Query: 265 IIGVDLARDILQSNPNNYAVVVSTE 289
IIGVDLARDILQSNPNNYAVVVSTE
Sbjct: 363 IIGVDLARDILQSNPNNYAVVVSTE 437
>TC9727 weakly similar to UP|Q9FV67 (Q9FV67) Fatty acid elongase 1-like
protein, partial (40%)
Length = 731
Score = 227 bits (579), Expect = 3e-60
Identities = 106/209 (50%), Positives = 159/209 (75%)
Frame = +2
Query: 37 RRLPDFLQSVNLKYVKLGYHYLINHGIYLFTIPLLLVVFSAEVGSLSKEDLWKKLWEDAS 96
R LPDF SV LKYVKLGYHYLI HG+YLF PL++++ +A++ + S +D++ +W++
Sbjct: 113 RNLPDFKNSVKLKYVKLGYHYLITHGMYLFLSPLVVLI-AAQLSTFSIKDIYD-IWDNLQ 286
Query: 97 YDLASVLASFAVFVFTLSVYFMSKPRPIYLIDFACYRPEDELKVSREQYIELASKSGKFD 156
Y+L SV+ + VF ++YF+++P+P+YL++F+CY+PE+ K +++++++ + SG F
Sbjct: 287 YNLISVIICSTLLVFLSTLYFLTRPKPVYLVNFSCYKPEEARKCTKKKFMDHSRMSGFFT 466
Query: 157 EESLAFQKRILMSSGIGDETYIPKAVVSSTTNTATMKEGRAEASTVMFGALDELFEKTGI 216
EE+L FQ++IL SG+G+ TY P+AV++ N TM E R EA VM+GA+DELF KT +
Sbjct: 467 EENLDFQRKILERSGLGENTYFPEAVLNIPPN-PTMHEARKEAEAVMYGAIDELFAKTSV 643
Query: 217 RPKDIGILVVNCSIFNPTPSLSAMIINHY 245
+ KDIGIL+VNCS+F PTPSLSAMI+NHY
Sbjct: 644 KAKDIGILIVNCSLFCPTPSLSAMIVNHY 730
>TC12061 similar to UP|Q9LN49 (Q9LN49) F18O14.21, partial (35%)
Length = 613
Score = 204 bits (519), Expect(2) = 2e-57
Identities = 98/165 (59%), Positives = 129/165 (77%)
Frame = +1
Query: 298 GKERSMLIPNCFFRMGCSALLLSNRRFDYNRAKYRLEHIVRTHKGADDRSFRCVYQEEDD 357
G ++SMLIPNC FR+G SA+LLSN+ D RAKY+L H+VRTHKGADD++FRCVYQE+D+
Sbjct: 7 GNKKSMLIPNCLFRVGGSAVLLSNKASDKFRAKYKLVHVVRTHKGADDKAFRCVYQEQDE 186
Query: 358 QKFKGLKISKDLIEIGGDALKTNITTLGPLVLPFSEQLLFFATLVWRHLFGGKSDGNSSP 417
G+ +SKDL+ I G ALKTNITTLGPLVLP SEQLLFFATL+ + +F K+
Sbjct: 187 AGKTGVSLSKDLMAIAGGALKTNITTLGPLVLPVSEQLLFFATLIVKKVFNAKA------ 348
Query: 418 SMKKPYIPNYKLAFEHFCVHAASKPILDELQRNLELSEKNMEASR 462
KPYIP++KLAF+HFC+HA + ++DEL++NL+L ++EAS+
Sbjct: 349 ---KPYIPDFKLAFDHFCIHAGGRAVIDELEKNLQLLPGHVEASK 474
Score = 35.4 bits (80), Expect(2) = 2e-57
Identities = 16/25 (64%), Positives = 18/25 (72%)
Frame = +3
Query: 469 GNTSSSSIWYELAYMEAKERVRRGD 493
GNTSSSSIWYEL Y EA + R +
Sbjct: 495 GNTSSSSIWYELGYTEAXGKGXRXE 569
>AV419456
Length = 372
Score = 207 bits (527), Expect = 3e-54
Identities = 104/104 (100%), Positives = 104/104 (100%)
Frame = +3
Query: 1 MARNSERDMFSAEIVNRGIESSGPNAGSLTFSVRVRRRLPDFLQSVNLKYVKLGYHYLIN 60
MARNSERDMFSAEIVNRGIESSGPNAGSLTFSVRVRRRLPDFLQSVNLKYVKLGYHYLIN
Sbjct: 60 MARNSERDMFSAEIVNRGIESSGPNAGSLTFSVRVRRRLPDFLQSVNLKYVKLGYHYLIN 239
Query: 61 HGIYLFTIPLLLVVFSAEVGSLSKEDLWKKLWEDASYDLASVLA 104
HGIYLFTIPLLLVVFSAEVGSLSKEDLWKKLWEDASYDLASVLA
Sbjct: 240 HGIYLFTIPLLLVVFSAEVGSLSKEDLWKKLWEDASYDLASVLA 371
>TC12004 similar to UP|Q8VWP9 (Q8VWP9) Fiddlehead-like protein, partial
(18%)
Length = 567
Score = 202 bits (514), Expect = 1e-52
Identities = 96/99 (96%), Positives = 98/99 (98%)
Frame = +3
Query: 440 SKPILDELQRNLELSEKNMEASRMTLHRFGNTSSSSIWYELAYMEAKERVRRGDRVWQLA 499
SKPILDELQRNLELSEKNMEASRMTLHRFGNTSSSSIWYELAYMEAKERVRRGDRVWQLA
Sbjct: 3 SKPILDELQRNLELSEKNMEASRMTLHRFGNTSSSSIWYELAYMEAKERVRRGDRVWQLA 182
Query: 500 FGSGFKCNSVVWLSMKRVNKPSRNNPWLDCINRYPVSLN 538
FGSGFKC+ VVWLSMKRVN+PSRNNPWLDCINRYPVSLN
Sbjct: 183 FGSGFKCHRVVWLSMKRVNQPSRNNPWLDCINRYPVSLN 299
>TC8766 similar to UP|Q8LAZ1 (Q8LAZ1) Very-long-chain fatty acid condensing
enzyme CUT1, partial (31%)
Length = 462
Score = 197 bits (500), Expect = 5e-51
Identities = 93/160 (58%), Positives = 125/160 (78%)
Frame = +3
Query: 279 PNNYAVVVSTEMVGFNWYQGKERSMLIPNCFFRMGCSALLLSNRRFDYNRAKYRLEHIVR 338
PN+ AVVVSTE++ N+YQG ER+ML+PNC FRMG +A++LSNR + +RAKY+L H+VR
Sbjct: 3 PNSNAVVVSTEIITPNYYQGNERAMLLPNCLFRMGGAAIMLSNRSSESSRAKYKLVHVVR 182
Query: 339 THKGADDRSFRCVYQEEDDQKFKGLKISKDLIEIGGDALKTNITTLGPLVLPFSEQLLFF 398
THKG+DD+++RCV++EED + G+ + KDL+ I G+ALK+NITT+GPLVLP SEQ+LF
Sbjct: 183 THKGSDDKAYRCVFEEEDKEGKVGISLQKDLMAIAGEALKSNITTMGPLVLPASEQVLFL 362
Query: 399 ATLVWRHLFGGKSDGNSSPSMKKPYIPNYKLAFEHFCVHA 438
ATL+ R +F K KPYIP++K AFEHFC+HA
Sbjct: 363 ATLIGRKIFNPK---------WKPYIPDFKQAFEHFCIHA 455
>BP036446
Length = 522
Score = 184 bits (466), Expect = 4e-47
Identities = 94/151 (62%), Positives = 113/151 (74%)
Frame = -1
Query: 377 LKTNITTLGPLVLPFSEQLLFFATLVWRHLFGGKSDGNSSPSMKKPYIPNYKLAFEHFCV 436
LK NITTLGPLVLP SEQ FF LV+R KPYIP+YKLAFEH CV
Sbjct: 522 LKDNITTLGPLVLPVSEQFHFFTNLVFRR------------KKTKPYIPDYKLAFEHVCV 379
Query: 437 HAASKPILDELQRNLELSEKNMEASRMTLHRFGNTSSSSIWYELAYMEAKERVRRGDRVW 496
A SK +LDE+Q+NLEL+E+ MEASR TL RFGNTSSSS WYELAY+E +ERV++GDRV
Sbjct: 378 VATSKKVLDEVQKNLELTEEYMEASRKTLERFGNTSSSSNWYELAYLEFEERVQKGDRVC 199
Query: 497 QLAFGSGFKCNSVVWLSMKRVNKPSRNNPWL 527
Q+A GSGF CNSVVW ++K V +P++ +PW+
Sbjct: 198 QIAAGSGFMCNSVVWKALKNVGRPNQ-SPWI 109
>AV407589
Length = 418
Score = 181 bits (460), Expect = 2e-46
Identities = 84/139 (60%), Positives = 112/139 (80%)
Frame = +1
Query: 113 LSVYFMSKPRPIYLIDFACYRPEDELKVSREQYIELASKSGKFDEESLAFQKRILMSSGI 172
+++YFMS+PR +YL+DF+CY+PE E V+RE +++ + +G F EE+LAFQK+IL SG+
Sbjct: 1 ITLYFMSRPRGVYLVDFSCYKPEPECNVTREIFMQRSELTGTFTEENLAFQKKILERSGL 180
Query: 173 GDETYIPKAVVSSTTNTATMKEGRAEASTVMFGALDELFEKTGIRPKDIGILVVNCSIFN 232
G +TY+P A++S N M E R EA VMFGA+D+L EKTG++ KDIGILVVNCS+FN
Sbjct: 181 GQKTYLPPAILSIPPNPC-MAEARKEAEQVMFGAIDQLLEKTGVKAKDIGILVVNCSLFN 357
Query: 233 PTPSLSAMIINHYKMRGNI 251
PTPSLSAMI+NHYK+RGNI
Sbjct: 358 PTPSLSAMIVNHYKLRGNI 414
>TC8454 similar to UP|Q84UX3 (Q84UX3) Fatty acid elongase (Fragment),
partial (50%)
Length = 661
Score = 178 bits (451), Expect = 2e-45
Identities = 76/115 (66%), Positives = 97/115 (84%)
Frame = +1
Query: 421 KPYIPNYKLAFEHFCVHAASKPILDELQRNLELSEKNMEASRMTLHRFGNTSSSSIWYEL 480
KPYIP++KLAFEHFC+HA + +LDEL++NL+LS+ +ME SRMTL+RFGNTSSSS+WYEL
Sbjct: 22 KPYIPDFKLAFEHFCIHAGGRAVLDELEKNLDLSDWHMEPSRMTLNRFGNTSSSSLWYEL 201
Query: 481 AYMEAKERVRRGDRVWQLAFGSGFKCNSVVWLSMKRVNKPSRNNPWLDCINRYPV 535
AY EAK R+R+GDR WQ+AFGSGFKCNS VW ++K +N NPW+D I+ +PV
Sbjct: 202 AYTEAKGRIRKGDRTWQIAFGSGFKCNSAVWQALKTINPAKEKNPWMDEIHEFPV 366
>AV426594
Length = 415
Score = 176 bits (447), Expect = 7e-45
Identities = 85/137 (62%), Positives = 106/137 (77%)
Frame = +1
Query: 261 CSAGIIGVDLARDILQSNPNNYAVVVSTEMVGFNWYQGKERSMLIPNCFFRMGCSALLLS 320
CSAGII +DLA+++LQ +PN+YA+VVS E + NWY G +RS L+ NC FRMG +A+LLS
Sbjct: 4 CSAGIISIDLAKELLQVHPNSYALVVSMENITLNWYPGNDRSKLVSNCLFRMGGAAILLS 183
Query: 321 NRRFDYNRAKYRLEHIVRTHKGADDRSFRCVYQEEDDQKFKGLKISKDLIEIGGDALKTN 380
N+ D R+KY+L VRTHKG+DD+ F CV QEED G+ +SKDL+ + GDALKTN
Sbjct: 184 NKSSDRRRSKYQLVDTVRTHKGSDDKCFSCVTQEEDANGKTGVTLSKDLMAVAGDALKTN 363
Query: 381 ITTLGPLVLPFSEQLLF 397
ITTLGPLVLP SEQLLF
Sbjct: 364 ITTLGPLVLPTSEQLLF 414
>AV409201
Length = 426
Score = 174 bits (442), Expect = 2e-44
Identities = 85/148 (57%), Positives = 113/148 (75%)
Frame = +2
Query: 283 AVVVSTEMVGFNWYQGKERSMLIPNCFFRMGCSALLLSNRRFDYNRAKYRLEHIVRTHKG 342
AVVVSTE++ N+YQG ER+ML+PNC FRMG +A+LL+NR+ RAKYRL H+VRTHKG
Sbjct: 8 AVVVSTEIITPNYYQGNERAMLLPNCLFRMGGAAILLTNRKSAKTRAKYRLVHVVRTHKG 187
Query: 343 ADDRSFRCVYQEEDDQKFKGLKISKDLIEIGGDALKTNITTLGPLVLPFSEQLLFFATLV 402
+DD+++RCVY+EED + G+ +SKDL+ I G+ALK+NITT+GP+VLP SEQL+F TL+
Sbjct: 188 SDDKAYRCVYEEEDKEGKVGISLSKDLMAIAGEALKSNITTIGPIVLPASEQLIFLVTLI 367
Query: 403 WRHLFGGKSDGNSSPSMKKPYIPNYKLA 430
R +F K KPYIP++K A
Sbjct: 368 GRKIFNPK---------WKPYIPDFKQA 424
>TC18581 similar to UP|Q9FG87 (Q9FG87) Beta-ketoacyl-CoA synthase
(AT5g43760/MQD19_11), partial (28%)
Length = 465
Score = 144 bits (363), Expect(2) = 1e-43
Identities = 69/107 (64%), Positives = 86/107 (79%)
Frame = +2
Query: 301 RSMLIPNCFFRMGCSALLLSNRRFDYNRAKYRLEHIVRTHKGADDRSFRCVYQEEDDQKF 360
R ML+ NC FRMG +A+LLS++ D RAKY+L H VRTHKGADD+S+ CV+QEED+ K
Sbjct: 122 RPMLVSNCLFRMGGAAILLSSKSSDRRRAKYQLVHTVRTHKGADDKSYGCVFQEEDETKT 301
Query: 361 KGLKISKDLIEIGGDALKTNITTLGPLVLPFSEQLLFFATLVWRHLF 407
G+ +SKDL+ + G+ALKTNITTLGPLVLP SEQ+LFF TLV R +F
Sbjct: 302 VGVALSKDLMAVAGEALKTNITTLGPLVLPMSEQILFFVTLVARKVF 442
Score = 49.7 bits (117), Expect(2) = 1e-43
Identities = 21/37 (56%), Positives = 29/37 (77%)
Frame = +1
Query: 262 SAGIIGVDLARDILQSNPNNYAVVVSTEMVGFNWYQG 298
SAG+I +DLA+ +LQ +PN+YA+VVS E + NWY G
Sbjct: 4 SAGLISIDLAKQLLQVHPNSYALVVSMENITLNWYFG 114
>TC8767 homologue to UP|Q8LAZ1 (Q8LAZ1) Very-long-chain fatty acid
condensing enzyme CUT1, partial (23%)
Length = 770
Score = 158 bits (399), Expect = 2e-39
Identities = 71/110 (64%), Positives = 90/110 (81%)
Frame = +1
Query: 428 KLAFEHFCVHAASKPILDELQRNLELSEKNMEASRMTLHRFGNTSSSSIWYELAYMEAKE 487
K AFEHFC+HA + ++DELQRNL+LS +++EASRMTLHRFGNTSSSS+WYEL Y+E+K
Sbjct: 1 KQAFEHFCIHAGGRAVIDELQRNLQLSAEHVEASRMTLHRFGNTSSSSLWYELNYIESKG 180
Query: 488 RVRRGDRVWQLAFGSGFKCNSVVWLSMKRVNKPSRNNPWLDCINRYPVSL 537
R++RGDRVWQ+AFGSGFKCNS VW + + P + P DCI+RYPV +
Sbjct: 181 RMKRGDRVWQIAFGSGFKCNSAVWKCNRTIKTPF-DGPCSDCIDRYPVHI 327
>AV420402
Length = 221
Score = 150 bits (380), Expect = 4e-37
Identities = 73/73 (100%), Positives = 73/73 (100%)
Frame = +3
Query: 312 MGCSALLLSNRRFDYNRAKYRLEHIVRTHKGADDRSFRCVYQEEDDQKFKGLKISKDLIE 371
MGCSALLLSNRRFDYNRAKYRLEHIVRTHKGADDRSFRCVYQEEDDQKFKGLKISKDLIE
Sbjct: 3 MGCSALLLSNRRFDYNRAKYRLEHIVRTHKGADDRSFRCVYQEEDDQKFKGLKISKDLIE 182
Query: 372 IGGDALKTNITTL 384
IGGDALKTNITTL
Sbjct: 183 IGGDALKTNITTL 221
>AV773511
Length = 463
Score = 138 bits (348), Expect = 2e-33
Identities = 62/98 (63%), Positives = 83/98 (84%)
Frame = -2
Query: 441 KPILDELQRNLELSEKNMEASRMTLHRFGNTSSSSIWYELAYMEAKERVRRGDRVWQLAF 500
+ ++DEL++NL+L ++EASRMTLHRFG TSSSSIWYELAY+EAK RVR+G+R+WQ+AF
Sbjct: 462 RAVIDELEKNLQLLPVHVEASRMTLHRFGYTSSSSIWYELAYIEAKGRVRKGNRLWQIAF 283
Query: 501 GSGFKCNSVVWLSMKRVNKPSRNNPWLDCINRYPVSLN 538
GSGFKCNS VW +++ V + S N PW DCI++YPV ++
Sbjct: 282 GSGFKCNSAVWEALRNV-RSSPNGPWEDCIDKYPVEID 172
>TC16819 similar to UP|Q8LAZ1 (Q8LAZ1) Very-long-chain fatty acid condensing
enzyme CUT1, partial (21%)
Length = 723
Score = 138 bits (347), Expect = 3e-33
Identities = 60/97 (61%), Positives = 80/97 (81%)
Frame = +1
Query: 441 KPILDELQRNLELSEKNMEASRMTLHRFGNTSSSSIWYELAYMEAKERVRRGDRVWQLAF 500
+ ++DELQ+NL+LS +++EASRMTLHRFGNTSSSS+WYEL Y+E+K R++RGDR+WQ+ F
Sbjct: 4 RAVIDELQKNLQLSAEHVEASRMTLHRFGNTSSSSLWYELNYIESKGRMKRGDRIWQIGF 183
Query: 501 GSGFKCNSVVWLSMKRVNKPSRNNPWLDCINRYPVSL 537
GSGFKCNS VW + + P + PW DCI+RYPV +
Sbjct: 184 GSGFKCNSAVWKCNRSIKTPV-DGPWEDCIDRYPVHI 291
>AV423524
Length = 528
Score = 137 bits (344), Expect = 6e-33
Identities = 65/156 (41%), Positives = 101/156 (64%)
Frame = +2
Query: 162 FQKRILMSSGIGDETYIPKAVVSSTTNTATMKEGRAEASTVMFGALDELFEKTGIRPKDI 221
F + ++SSGIG++TY P+ V + T+++G E ++ +L ++T I P +I
Sbjct: 59 FLLKAIVSSGIGEQTYAPRNVFDGREASPTLEDGILEMEEFFNDSIAKLLQRTSISPSEI 238
Query: 222 GILVVNCSIFNPTPSLSAMIINHYKMRGNILSYNLGGMGCSAGIIGVDLARDILQSNPNN 281
+LVVN S+F PSLS+ IIN YK+R +I YNL GMGCSA +I VD+ ++I ++ N
Sbjct: 239 DVLVVNISMFAAVPSLSSRIINRYKLRHDIKVYNLTGMGCSASLISVDIVQNIFKTQKNK 418
Query: 282 YAVVVSTEMVGFNWYQGKERSMLIPNCFFRMGCSAL 317
A++V++E + NWY G +RSM++ NC FR G A+
Sbjct: 419 LALLVTSESLSPNWYNGNDRSMILANCLFRSGGCAM 526
>AV427430
Length = 345
Score = 135 bits (339), Expect = 2e-32
Identities = 61/113 (53%), Positives = 87/113 (76%)
Frame = +2
Query: 262 SAGIIGVDLARDILQSNPNNYAVVVSTEMVGFNWYQGKERSMLIPNCFFRMGCSALLLSN 321
SAG+I +DLA+D+L++NPN+YAVVVSTE + NWY G +RSML+ NC FRMG +A+LLSN
Sbjct: 5 SAGLISIDLAKDLLKANPNSYAVVVSTENITLNWYFGNDRSMLLCNCIFRMGGAAVLLSN 184
Query: 322 RRFDYNRAKYRLEHIVRTHKGADDRSFRCVYQEEDDQKFKGLKISKDLIEIGG 374
+ D ++KY L H VRTHKG DD+ + CVYQ ED++ G+ ++++L+ + G
Sbjct: 185 KSSDRAKSKYELVHTVRTHKGFDDKHYNCVYQMEDEKGKVGVCLARELMAVAG 343
>TC19140 similar to PIR|F86141|F86141 protein T25K16.11 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(14%)
Length = 525
Score = 131 bits (329), Expect = 3e-31
Identities = 63/96 (65%), Positives = 78/96 (80%), Gaps = 2/96 (2%)
Frame = +2
Query: 441 KPILDELQRNLELSEKNMEASRMTLHRFGNTSSSSIWYELAYMEAKERVRRGDRVWQLAF 500
+ +LDE+Q+NLELSE +ME SRMTLHRFGNTSSSS+WYELAY EAK RV +GDRV Q+AF
Sbjct: 2 RAVLDEMQKNLELSEWHMEPSRMTLHRFGNTSSSSLWYELAYTEAKARVGKGDRV*QIAF 181
Query: 501 GSGFKCNSVVWLSMKRVNKPS--RNNPWLDCINRYP 534
GSGFKCNS VW +++ + + R NPW D I++YP
Sbjct: 182 GSGFKCNSAVWKAVRDMPELGDWRGNPWDDSIDKYP 289
>AV778558
Length = 444
Score = 130 bits (326), Expect = 7e-31
Identities = 62/151 (41%), Positives = 99/151 (65%)
Frame = -2
Query: 385 GPLVLPFSEQLLFFATLVWRHLFGGKSDGNSSPSMKKPYIPNYKLAFEHFCVHAASKPIL 444
G +LP SE+ + +++ + KS+G Y+PN+K +HFC+ + + ++
Sbjct: 443 GSEILPPSEKFWYGVSVIQKKFI--KSEGI--------YVPNFKTVIQHFCLPCSGRAVI 294
Query: 445 DELQRNLELSEKNMEASRMTLHRFGNTSSSSIWYELAYMEAKERVRRGDRVWQLAFGSGF 504
E+ + L+L+E+++E + MTL+RFG+ SSSS+WYELAY+EAKERV++GD+VW L GSG
Sbjct: 293 REVGKGLKLAERDIEPAMMTLYRFGDPSSSSLWYELAYLEAKERVQKGDKVWPLGMGSGP 114
Query: 505 KCNSVVWLSMKRVNKPSRNNPWLDCINRYPV 535
KC VV ++ + S+ PW DCI+ YP+
Sbjct: 113 KCIRVVLKCVRPLVGESQKGPWADCIHPYPI 21
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.137 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,752,702
Number of Sequences: 28460
Number of extensions: 142647
Number of successful extensions: 712
Number of sequences better than 10.0: 71
Number of HSP's better than 10.0 without gapping: 694
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 696
length of query: 538
length of database: 4,897,600
effective HSP length: 95
effective length of query: 443
effective length of database: 2,193,900
effective search space: 971897700
effective search space used: 971897700
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0133.9


