

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0128.10
(939 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
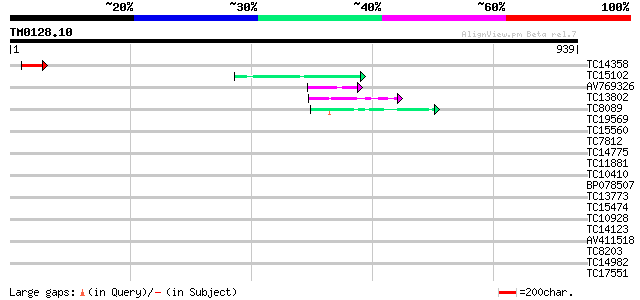
Score E
Sequences producing significant alignments: (bits) Value
TC14358 89 3e-18
TC15102 similar to UP|Q7X9B3 (Q7X9B3) 9/13 hydroperoxide lyase, ... 45 4e-05
AV769326 45 5e-05
TC13802 similar to UP|Q9M1T1 (Q9M1T1) Sugar-phosphate isomerase-... 43 2e-04
TC8089 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabinog... 43 3e-04
TC19569 similar to UP|AAR11301 (AAR11301) Lectin-like receptor k... 40 0.002
TC15560 similar to SP|Q03173|NDPP_MOUSE NPC derived proline rich... 39 0.003
TC7812 UP|Q9LKJ6 (Q9LKJ6) Water-selective transport intrinsic me... 39 0.003
TC14775 PIR|S14181|S14181 DNA-directed RNA polymerase largest c... 39 0.003
TC11881 similar to PIR|T48303|T48303 meiosis specific-like prote... 38 0.006
TC10410 weakly similar to UP|Q8H5W8 (Q8H5W8) OJ1123_B01.10 prote... 38 0.006
BP078507 38 0.008
TC13773 similar to UP|AAR20741 (AAR20741) At4g22560, partial (11%) 38 0.008
TC15474 UP|Q7Y1B9 (Q7Y1B9) Ammonium transporter, complete 37 0.011
TC10928 weakly similar to PIR|G96668|G96668 protein F1N19.7 [imp... 37 0.014
TC14123 similar to UP|Q93X25 (Q93X25) 2-Cys peroxiredoxin, parti... 37 0.019
AV411518 36 0.025
TC8203 similar to UP|Q8L9C3 (Q8L9C3) Copia-like retroelement pol... 35 0.042
TC14982 similar to UP|Q9FPI2 (Q9FPI2) At1g17620, partial (32%) 35 0.042
TC17551 weakly similar to UP|AAR24662 (AAR24662) At5g26960, part... 35 0.042
>TC14358
Length = 1084
Score = 89.0 bits (219), Expect = 3e-18
Identities = 41/43 (95%), Positives = 43/43 (99%)
Frame = -3
Query: 20 RIRTCLSKHSRQTLEYHSCSSTNKYLLSSKHRVVPHDQILILQ 62
RIRTCLSKHSRQTLEYH+CSSTNKYLLSSKH+VVPHDQILILQ
Sbjct: 197 RIRTCLSKHSRQTLEYHNCSSTNKYLLSSKHKVVPHDQILILQ 69
Score = 27.7 bits (60), Expect = 8.8
Identities = 13/21 (61%), Positives = 16/21 (75%), Gaps = 1/21 (4%)
Frame = -3
Query: 2 TVGGHFWNFQNLLWLN-LGRI 21
TVGG NFQ LLWL+ LG++
Sbjct: 611 TVGGQICNFQILLWLDILGKV 549
>TC15102 similar to UP|Q7X9B3 (Q7X9B3) 9/13 hydroperoxide lyase, partial
(68%)
Length = 1097
Score = 45.4 bits (106), Expect = 4e-05
Identities = 55/223 (24%), Positives = 85/223 (37%), Gaps = 7/223 (3%)
Frame = +1
Query: 373 PPAPEFPSPPKKKAQRKMIPEESSEESDVPLVKKKKRKPDDGDDSDNEDGPPKKKKKQVR 432
PP +PP + + P P K + P K +
Sbjct: 223 PPESRSTTPPSSEPTCLLAPAS-------PPTPKSSHSSTPPPSRSSSTTPKSKNATSLT 381
Query: 433 IVVKPTRVEPAAEVVRRTE-PPARVTRSSAHSS-KSALASDDDLNLFDALPISAMLKHSS 490
P PAA ++ PP T SS SS KS+L + +L A P ++
Sbjct: 382 APSCPPPASPAATAPAPSKTPPNPPTNSSKPSSCKSSLPNTTPSSLSSAPP-----SPTT 546
Query: 491 NPLTT--IPESQPAAQTTSPSHSPRS-SFFQPSPTEAPLWNLLQNQPSRSEEPTSPITIQ 547
+P++T PE+Q +T PS PRS S SP P + S P S +++
Sbjct: 547 SPISTPSSPENQEKPASTPPSAPPRSTSSSSSSPINTPRKPSSATKAPASSPPGSELSLH 726
Query: 548 YNPLSSEPIIHAQPEPTHTEP--EPRTSDHSVPRASERLALKP 588
++P S+ P+ + +EP R + R S R++L P
Sbjct: 727 HSPPSASRKSSTTPKISSSEPSHSRRGQ*NGATRNSARVSLPP 855
Score = 37.4 bits (85), Expect = 0.011
Identities = 59/267 (22%), Positives = 86/267 (32%), Gaps = 22/267 (8%)
Frame = +1
Query: 482 ISAMLKHSSNPLTTIP----------ESQPAAQTTSPSHSPRSSFFQPSPTEAPLWNLLQ 531
+ A SS P TT S P +++T+P S + P+ P +
Sbjct: 139 LEATASPSSEPCTTATTTSTTKAATNSSPPESRSTTPPSSEPTCLLAPASPPTPKSSHSS 318
Query: 532 NQPSRSEEPTSP-------ITIQYNPLSSEPIIHAQPEPTHTEPEPRTSDHSVPRASERL 584
P T+P +T P + P A P P+ T P P T+ L
Sbjct: 319 TPPPSRSSSTTPKSKNATSLTAPSCPPPASPAATA-PAPSKTPPNPPTNSSKPSSCKSSL 495
Query: 585 ALKPTDTDSSTAFTPVSFPTNVADSSPSNNSESIRKFMEVRKEKVSTLEEYYLTCPSPRR 644
P T SS + P S PT S+PS+ P +
Sbjct: 496 ---PNTTPSSLSSAPPS-PTTSPISTPSS--------------------------PENQE 585
Query: 645 YPGPRPERLVDPDEPILANPLQEADPLAQQAHPAPDQQEPIQPEPEPEQSVSNQ----SS 700
P P P P + + P+ P+ + P P E S+ + S
Sbjct: 586 KPASTP-----PSAPPRSTSSSSSSPINTPRKPSSATKAPASSPPGSELSLHHSPPSASR 750
Query: 701 VRSPHPLVETSEP-HLGTSKPNAQTFN 726
S P + +SEP H + N T N
Sbjct: 751 KSSTTPKISSSEPSHSRRGQ*NGATRN 831
>AV769326
Length = 262
Score = 45.1 bits (105), Expect = 5e-05
Identities = 30/91 (32%), Positives = 43/91 (46%)
Frame = +2
Query: 493 LTTIPESQPAAQTTSPSHSPRSSFFQPSPTEAPLWNLLQNQPSRSEEPTSPITIQYNPLS 552
+++ P PA T+SPS P ++ PSP P + N PS + +P SP L+
Sbjct: 2 ISSAPRCFPATTTSSPSSPPSTTPTTPSPPSDP-HSPPSNPPSAASDPNSP------SLT 160
Query: 553 SEPIIHAQPEPTHTEPEPRTSDHSVPRASER 583
+ + PT TEP +S S P AS R
Sbjct: 161 APSLQKPPSSPTSTEPPSSSSTPSAPSASTR 253
>TC13802 similar to UP|Q9M1T1 (Q9M1T1) Sugar-phosphate isomerase-like
protein, partial (41%)
Length = 522
Score = 43.1 bits (100), Expect = 2e-04
Identities = 45/160 (28%), Positives = 67/160 (41%), Gaps = 4/160 (2%)
Frame = +2
Query: 495 TIPESQPAA---QTTSPSHSPRSSFFQPSPTEAPLWNLLQNQPSRSEEPTSPITI-QYNP 550
T P SQ ++ +TTS + SP S+ +PSP+ AP L PS S SP + +P
Sbjct: 77 TKPPSQISSNPNKTTSTTSSPTSTTPKPSPSPAPSSTL--RAPSSSPASASPASSPTKSP 250
Query: 551 LSSEPIIHAQPEPTHTEPEPRTSDHSVPRASERLALKPTDTDSSTAFTPVSFPTNVADSS 610
S P A P T P TS+ S PT T SS++ +P +
Sbjct: 251 KPSSPSASAPPSSTPLTPSTATSESS-----------PTATSSSSSASP----------A 367
Query: 611 PSNNSESIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRP 650
P + ++ + +E++ + PSP P P P
Sbjct: 368 PPTSFSALSRALELKA---------LASSPSPPSSPPPSP 460
>TC8089 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabinogalactan
protein 1 precursor, partial (43%)
Length = 890
Score = 42.7 bits (99), Expect = 3e-04
Identities = 60/229 (26%), Positives = 83/229 (36%), Gaps = 15/229 (6%)
Frame = +2
Query: 499 SQPAAQTTSPSHS--PRSSFFQPSPTE---APLW-----NLLQNQPSRSEEPTSPITIQY 548
S P + TTSPS + PRS+ SP+ P W N Q+ PSR+ P++ +
Sbjct: 263 SSPPSTTTSPSPTSPPRSTNGPQSPSAPSTTPPWTTSSQNTHQSPPSRTSSPSTSSSTTS 442
Query: 549 NPLSSEPIIHAQPEPTHTEPEPRTSDHSVPRASERLALKPTDTDSSTAFTPVSFPTNVAD 608
P SS A P P P + PRA + PT S A S P
Sbjct: 443 APRSSTRSPTAPPSPPQCTKPP-----APPRAPPDSSTSPT----SAAGKSDSAPRTTTA 595
Query: 609 SSPSNNSESIRKFMEVRKEKVSTLEEYYLTCPSPRRYPGPRPERLVDP--DEPILANPLQ 666
SP +S RK SP P R R P +P+ +PL
Sbjct: 596 PSPLRSSNP*RK--------------------SPTTSPSSRSVRFFPPPQQKPLHPHPLS 715
Query: 667 EADP---LAQQAHPAPDQQEPIQPEPEPEQSVSNQSSVRSPHPLVETSE 712
P + A +P P + +P + S+V P P T++
Sbjct: 716 RISPPLCPSTVARSSPTLSPPRRTRTQPSPTT---STVG*PFPAPSTTQ 853
Score = 36.2 bits (82), Expect = 0.025
Identities = 41/126 (32%), Positives = 49/126 (38%), Gaps = 2/126 (1%)
Frame = +2
Query: 450 TEPPA--RVTRSSAHSSKSALASDDDLNLFDALPISAMLKHSSNPLTTIPESQPAAQTTS 507
T+PPA R S+ S SA D P SSNP P TTS
Sbjct: 497 TKPPAPPRAPPDSSTSPTSAAGKSDSAPRTTTAPSPL---RSSNP*RKSP-------TTS 646
Query: 508 PSHSPRSSFFQPSPTEAPLWNLLQNQPSRSEEPTSPITIQYNPLSSEPIIHAQPEPTHTE 567
P S RS F P P + PL + SR P P T+ + + P P T T+
Sbjct: 647 P--SSRSVRFFPPPQQKPLH---PHPLSRISPPLCPSTVARSSPTLSP-----PRRTRTQ 796
Query: 568 PEPRTS 573
P P TS
Sbjct: 797 PSPTTS 814
>TC19569 similar to UP|AAR11301 (AAR11301) Lectin-like receptor kinase 1;1,
partial (16%)
Length = 486
Score = 39.7 bits (91), Expect = 0.002
Identities = 43/142 (30%), Positives = 62/142 (43%), Gaps = 2/142 (1%)
Frame = +1
Query: 481 PISAMLKHSSNPLTTIPESQPAAQTTSPSHSPRSSFFQPSPTEAPLWNLLQNQPSRSEE- 539
P +L SS+ ++ P +Q P+HSP +S P+ T P P++ E
Sbjct: 22 PCGLLLHTSSSSSSSYSPPPPPSQ--QPTHSPSTS---PTSTTTP-----PPSPTKVTEN 171
Query: 540 -PTSPITIQYNPLSSEPIIHAQPEPTHTEPEPRTSDHSVPRASERLALKPTDTDSSTAFT 598
PT+P T +P SS A EP + P P TS P+ S PT +S + +
Sbjct: 172 PPTAPSTSTKSPTSS-----ASEEP--SPPNPSTSTTQQPKHS------PTSPPASPSPS 312
Query: 599 PVSFPTNVADSSPSNNSESIRK 620
P S A +SPS + S K
Sbjct: 313 PPSTKPPTAMASPSTSPLSATK 378
Score = 33.1 bits (74), Expect = 0.21
Identities = 41/161 (25%), Positives = 58/161 (35%), Gaps = 4/161 (2%)
Frame = +1
Query: 417 SDNEDGPPKKKKKQVRIVVKPTRVEPAAEVVRRTEPPARVTRSSAHSSKSALASDDDLNL 476
S + PP +Q PT P+ T PP T+ + + + S
Sbjct: 55 SSSSYSPPPPPSQQ------PTH-SPSTSPTSTTTPPPSPTKVTENPPTAPSTSTKS--- 204
Query: 477 FDALPISAMLKHSSNPLTTIPESQPAAQTTS-PSHSPRSSFFQPSPTEAPLWNLLQNQPS 535
P S+ + S P P+ TT P HSP S PSP+ P
Sbjct: 205 ----PTSSASEEPSPP-------NPSTSTTQQPKHSPTSPPASPSPS-----------PP 318
Query: 536 RSEEPTSPITIQYNPLS---SEPIIHAQPEPTHTEPEPRTS 573
++ PT+ + +PLS S P A P + T P TS
Sbjct: 319 STKPPTAMASPSTSPLSATKSHPTPPAAPSASSTPPPTTTS 441
>TC15560 similar to SP|Q03173|NDPP_MOUSE NPC derived proline rich protein 1
(NDPP-1). {Mus musculus;}, partial (8%)
Length = 1134
Score = 39.3 bits (90), Expect = 0.003
Identities = 41/138 (29%), Positives = 63/138 (44%), Gaps = 8/138 (5%)
Frame = +3
Query: 488 HSSNPLTTI------PESQPAAQTTSPSHSPRSSFFQPSPTEAPLWNLLQN-QPSRSEEP 540
H PLTT S P++ SPS P S P P PL + L + +PS + P
Sbjct: 99 HHHYPLTTTASASSSSSSSPSSS*PSPSLPPPSPPPLPPPQAPPLPSHLPSPEPSSTMPP 278
Query: 541 TSPITIQYNPLSSEPIIHAQPEPTHTEPEPRTSDHSVPRASERLALKPTDTDSSTAFTPV 600
++P P ++P P+ T ++P P ++ H VP +S PT S T +
Sbjct: 279 STP-----PPQPTKPC--PLPKSTPSQP-PSSAPHRVPTSSS--LASPTSPSSGPHSTTM 428
Query: 601 SFP-TNVADSSPSNNSES 617
+ P ++ S+PS NS +
Sbjct: 429 AAPFSSTRTSTPSPNSNN 482
Score = 35.8 bits (81), Expect = 0.032
Identities = 27/87 (31%), Positives = 38/87 (43%), Gaps = 5/87 (5%)
Frame = +3
Query: 489 SSNPLTTIPESQPAAQTTSPSHSPRSSFFQPSPTEAPLWNLLQNQPSRSE--EPTSPITI 546
S P +T+P S P Q T P P+S+ QP P+ AP P+ S PTSP +
Sbjct: 249 SPEPSSTMPPSTPPPQPTKPCPLPKSTPSQP-PSSAP-----HRVPTSSSLASPTSPSSG 410
Query: 547 QYNPLSSEPIIHAQ---PEPTHTEPEP 570
++ + P + P P P P
Sbjct: 411 PHSTTMAAPFSSTRTSTPSPNSNNPTP 491
>TC7812 UP|Q9LKJ6 (Q9LKJ6) Water-selective transport intrinsic membrane
protein 1, complete
Length = 1149
Score = 39.3 bits (90), Expect = 0.003
Identities = 39/126 (30%), Positives = 51/126 (39%), Gaps = 2/126 (1%)
Frame = +1
Query: 490 SNPLTTIPESQPAAQTTSPSHSPRSSFFQPSPT-EAPLWNLLQNQPSRSEEPTSPITIQY 548
S PL P P Q S SP SS P+P E+P N P + P + +
Sbjct: 97 SVPLRRQPTQTP*GQPWLSS-SPPSSSSSPAPVPESPTTNSPTTAPPPPPASSPPPSPTH 273
Query: 549 NPLSS-EPIIHAQPEPTHTEPEPRTSDHSVPRASERLALKPTDTDSSTAFTPVSFPTNVA 607
+P SS P P T T P P S +V P+ SST+ S P++
Sbjct: 274 SPSSSPSPSAPTSPAATSTPPSPSASSSAVT--------SPSSAVSSTSSLSFSDPSSPP 429
Query: 608 DSSPSN 613
SSPS+
Sbjct: 430 CSSPSS 447
>TC14775 PIR|S14181|S14181 DNA-directed RNA polymerase largest chain
(isoform B1) - soybean (fragment) {Glycine
max;} , partial (22%)
Length = 848
Score = 39.3 bits (90), Expect = 0.003
Identities = 33/123 (26%), Positives = 54/123 (43%), Gaps = 5/123 (4%)
Frame = +1
Query: 499 SQPAAQTTSPSHSPRSSFFQP-SPTEAPLWNLLQNQPSRSEEPTSPITIQYNPLSSEPII 557
+ P TSPS+SP S + P SP+ P + PS + P+SP +P S
Sbjct: 37 TSPGYSPTSPSYSPTSPSYSPTSPSYNP--QSAKYSPSLAYSPSSPRLSPSSPYSPTSPN 210
Query: 558 HAQPEPTHTEPEPRTSDHS---VPRASERLALKPTDTDSSTAFTP-VSFPTNVADSSPSN 613
++ P+++ P S S P + + P + SS F+P + + SPS+
Sbjct: 211 YSPTSPSYSPTSPSYSPSSPTYSPSSPYNSGVSPDYSPSSPQFSPSTGYSPSQPGYSPSS 390
Query: 614 NSE 616
S+
Sbjct: 391 TSQ 399
Score = 38.9 bits (89), Expect = 0.004
Identities = 38/120 (31%), Positives = 57/120 (46%), Gaps = 3/120 (2%)
Frame = +1
Query: 501 PAAQTTSPSHSPRSSFFQP-SPTEAPLWNLLQNQPSRSEEPTSPITIQYNPLSSEPIIHA 559
P+ TSP++SP S + P SP+ +P S S PTSP YNP S++
Sbjct: 1 PSYSPTSPAYSPTSPGYSPTSPSYSP--------TSPSYSPTSP---SYNPQSAK----Y 135
Query: 560 QPEPTHTEPEPRTSDHS--VPRASERLALKPTDTDSSTAFTPVSFPTNVADSSPSNNSES 617
P ++ PR S S P + P+ + +S +++P S PT + SSP N+ S
Sbjct: 136 SPSLAYSPSSPRLSPSSPYSPTSPNYSPTSPSYSPTSPSYSP-SSPT-YSPSSPYNSGVS 309
Score = 30.4 bits (67), Expect = 1.4
Identities = 28/104 (26%), Positives = 42/104 (39%), Gaps = 1/104 (0%)
Frame = +1
Query: 481 PISAMLKHSSNPLTTIPESQPAAQTTSPSHSPRSSFFQP-SPTEAPLWNLLQNQPSRSEE 539
P S L SS T P P TSPS+SP S + P SPT +P S
Sbjct: 157 PSSPRLSPSSPYSPTSPNYSP----TSPSYSPTSPSYSPSSPTYSP-----------SSP 291
Query: 540 PTSPITIQYNPLSSEPIIHAQPEPTHTEPEPRTSDHSVPRASER 583
S ++ Y+P S + P+ P ++ P+ +++
Sbjct: 292 YNSGVSPDYSPSSPQFSPSTGYSPSQPGYSPSSTSQYTPQTNDK 423
>TC11881 similar to PIR|T48303|T48303 meiosis specific-like protein -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(34%)
Length = 631
Score = 38.1 bits (87), Expect = 0.006
Identities = 38/127 (29%), Positives = 54/127 (41%), Gaps = 1/127 (0%)
Frame = +3
Query: 497 PESQPAAQTTSPSHSPRSSF-FQPSPTEAPLWNLLQNQPSRSEEPTSPITIQYNPLSSEP 555
P+++P TSPS S + + PSP P +L P+RS P+SP+T +P S P
Sbjct: 204 PQTKP*PSKTSPSPSAAARWRTSPSPPCNPTSSL---SPARSRTPSSPVTA--SPSMSPP 368
Query: 556 IIHAQPEPTHTEPEPRTSDHSVPRASERLALKPTDTDSSTAFTPVSFPTNVADSSPSNNS 615
A P + S + S RL P P + P + SS S+ S
Sbjct: 369 ---APPPTSSMSLNSTASSSRINPPSARLVTSP----------PSANPPSPPASSSSSTS 509
Query: 616 ESIRKFM 622
+ R FM
Sbjct: 510 SASRGFM 530
>TC10410 weakly similar to UP|Q8H5W8 (Q8H5W8) OJ1123_B01.10 protein, partial
(12%)
Length = 431
Score = 38.1 bits (87), Expect = 0.006
Identities = 43/144 (29%), Positives = 60/144 (40%), Gaps = 8/144 (5%)
Frame = +3
Query: 483 SAMLKHSSNPLTTIPESQP-------AAQTTSPSHSPRSSFFQPSPTEAPLWNLLQNQPS 535
+A + +S P + P QP ++ TT P +P P+PT P N N
Sbjct: 33 AATARSTSEPSQSQPPLQPPSSLPQTSSSTTPPISAPPHPPQNPNPTPIPAPNPNPN--- 203
Query: 536 RSEEPTSPITIQYNPLSSEPIIHAQPEPTHTEPEPRTSDHSVPRASERLALKPTDTDSST 595
PI IQ NP PI + P PT T P+PR + RA +P+ +
Sbjct: 204 -------PIQIQ-NP---NPIHNPNPRPTPTPPQPRPPP-TFNRAPPPQQQQPSHFSHFS 347
Query: 596 AFTPVSFPT-NVADSSPSNNSESI 618
+ P S P+ + A S S S SI
Sbjct: 348 SLPPSSAPSPSPASSFNSTPSPSI 419
>BP078507
Length = 414
Score = 37.7 bits (86), Expect = 0.008
Identities = 30/94 (31%), Positives = 37/94 (38%)
Frame = -1
Query: 488 HSSNPLTTIPESQPAAQTTSPSHSPRSSFFQPSPTEAPLWNLLQNQPSRSEEPTSPITIQ 547
H S+P P S + T SP S P+P +P QPS + PTSP
Sbjct: 372 HQSHPTQDTPSSHT*SSPTDKQSSPES---YPTPLHSP-------QPSPNA-PTSPYPAP 226
Query: 548 YNPLSSEPIIHAQPEPTHTEPEPRTSDHSVPRAS 581
+ L P QP PT +P T S P S
Sbjct: 225 SSQLRPPPSTSHQPPPTLLQPTSDTPSPSPPPPS 124
Score = 33.1 bits (74), Expect = 0.21
Identities = 28/95 (29%), Positives = 36/95 (37%)
Frame = -1
Query: 627 EKVSTLEEYYLTCPSPRRYPGPRPERLVDPDEPILANPLQEADPLAQQAHPAPDQQEPIQ 686
+K S+ E Y P+P P P P P P A Q P + P P +P
Sbjct: 315 DKQSSPESY----PTPLHSPQPSPNA---PTSPYPAPSSQLRPPPSTSHQPPPTLLQPTS 157
Query: 687 PEPEPEQSVSNQSSVRSPHPLVETSEPHLGTSKPN 721
P P S SS P P + PH +S P+
Sbjct: 156 DTPSP--SPPPPSSPNPPPPHHSPNPPHPPSSSPS 58
>TC13773 similar to UP|AAR20741 (AAR20741) At4g22560, partial (11%)
Length = 441
Score = 37.7 bits (86), Expect = 0.008
Identities = 27/91 (29%), Positives = 40/91 (43%)
Frame = +2
Query: 481 PISAMLKHSSNPLTTIPESQPAAQTTSPSHSPRSSFFQPSPTEAPLWNLLQNQPSRSEEP 540
P+ N L+ +P ++ A +T+ S S PSP +P + Q +++P
Sbjct: 182 PLKMSTTQIPNTLSPVPVAERAFKTSKTLFSTPPS---PSPNPSPKNHSRNTQVPDTQQP 352
Query: 541 TSPITIQYNPLSSEPIIHAQPEPTHTEPEPR 571
P I PLS+ P AQP PT P R
Sbjct: 353 LPPNLISKQPLSTAP--GAQPPPTPAMPRRR 439
>TC15474 UP|Q7Y1B9 (Q7Y1B9) Ammonium transporter, complete
Length = 1950
Score = 37.4 bits (85), Expect = 0.011
Identities = 34/134 (25%), Positives = 54/134 (39%), Gaps = 2/134 (1%)
Frame = -1
Query: 486 LKHSSNPLTTIPESQPAAQTTSPSHSPRSSFFQPSPTEAPLWNLLQNQPSRSEEPTSPIT 545
L H + T S P+ Q+ S + S ++ T AP + P S+ P P T
Sbjct: 1503 LHHGKHKQTHHASSNPSPQSHHLQKSSKVSSYEAHRTTAPSSP*PTHTPPISQSPAPPAT 1324
Query: 546 IQYNPLSSEPIIHAQPEPTHTEPEPRTSDHSVP--RASERLALKPTDTDSSTAFTPVSFP 603
+PL++ +P P +T+ P R + R A P + D T+F+P
Sbjct: 1323 SLRHPLTTRTASQRCRKPPPRTPPSQTAP*IKPPTRRTRREAAPPPE-DRRTSFSPQPN* 1147
Query: 604 TNVADSSPSNNSES 617
A + P N +S
Sbjct: 1146 ARSAPTLPQNRRQS 1105
>TC10928 weakly similar to PIR|G96668|G96668 protein F1N19.7 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(29%)
Length = 590
Score = 37.0 bits (84), Expect = 0.014
Identities = 29/99 (29%), Positives = 41/99 (41%), Gaps = 4/99 (4%)
Frame = -3
Query: 479 ALPISAMLKHS----SNPLTTIPESQPAAQTTSPSHSPRSSFFQPSPTEAPLWNLLQNQP 534
+LP+S + HS ++P + P + P TSPS +P ++ P P P P
Sbjct: 399 SLPLSPHVSHSRSDPTDPTHSAPPTPPTRPNTSPSQTPSAAAPPPPPPPPPA------AP 238
Query: 535 SRSEEPTSPITIQYNPLSSEPIIHAQPEPTHTEPEPRTS 573
S E S P P ++A P P PRTS
Sbjct: 237 SPLSETRSGAPRHARP----PPLYAPPAPPSPGCNPRTS 133
>TC14123 similar to UP|Q93X25 (Q93X25) 2-Cys peroxiredoxin, partial (89%)
Length = 1130
Score = 36.6 bits (83), Expect = 0.019
Identities = 22/52 (42%), Positives = 31/52 (59%), Gaps = 1/52 (1%)
Frame = +2
Query: 476 LFDALPISAM-LKHSSNPLTTIPESQPAAQTTSPSHSPRSSFFQPSPTEAPL 526
LF P +++ + +S NPL+T P S P+ T PSHS R SF + +E PL
Sbjct: 110 LFSPKPTTSLSIPNSLNPLSTKPFSLPSVSLTRPSHS-RRSFLVRATSELPL 262
>AV411518
Length = 360
Score = 36.2 bits (82), Expect = 0.025
Identities = 33/111 (29%), Positives = 44/111 (38%), Gaps = 10/111 (9%)
Frame = +3
Query: 505 TTSPSHSPRSSFFQPSPTEAPLWNLLQNQPSRSEEPT-----SPITIQYNPLSSEPIIHA 559
T S S SP +S+ PSP P PS PT S + +P + A
Sbjct: 6 TPSLSPSPATSWLPPSP*AHPTPATSWLPPSP*AHPTPAWAGSALASWASPCAPTSSGPA 185
Query: 560 QPEPTHTEPEPR-----TSDHSVPRASERLALKPTDTDSSTAFTPVSFPTN 605
P P+ T P PR TS + P PT + S+A P S P++
Sbjct: 186 SPSPSSTAPSPRPNPSSTSGPTSPTPHTPSPPPPTSSSPSSAIPPTSAPSS 338
>TC8203 similar to UP|Q8L9C3 (Q8L9C3) Copia-like retroelement pol
polyprotein, partial (96%)
Length = 1198
Score = 35.4 bits (80), Expect = 0.042
Identities = 30/110 (27%), Positives = 50/110 (45%), Gaps = 10/110 (9%)
Frame = +2
Query: 807 QVPIMMQLLYAEGS----QSIEAAKQRFARRVALHEQ---EQRHKLLEAIEEAKRKKAQA 859
Q+ I Q++ EG+ ++++A + + A+ + + K ++ I E Q
Sbjct: 461 QLRIHDQMIMLEGAKATTETVDALRTGASAMKAMQKATNIDDVDKTMDEINEQTENMKQI 640
Query: 860 EEAVRQ---AAAQVEQARLEAERLEVEAEALRCQALAPVVFTPAASASIP 906
+EA+ AAA ++ LEAE E+E L Q L P PAA P
Sbjct: 641 QEALSAPIGAAADFDEDELEAELEELEGAELEEQLLQPATTAPAAPVHAP 790
>TC14982 similar to UP|Q9FPI2 (Q9FPI2) At1g17620, partial (32%)
Length = 1036
Score = 35.4 bits (80), Expect = 0.042
Identities = 31/127 (24%), Positives = 50/127 (38%)
Frame = +3
Query: 435 VKPTRVEPAAEVVRRTEPPARVTRSSAHSSKSALASDDDLNLFDALPISAMLKHSSNPLT 494
+ P+ PAA + + SS+ SS + LAS P ++ ++ P
Sbjct: 153 LNPSTAAPAAAASAPSASG*SSSSSSSSSSSAVLAS----------PSTSSTAPTTPPSP 302
Query: 495 TIPESQPAAQTTSPSHSPRSSFFQPSPTEAPLWNLLQNQPSRSEEPTSPITIQYNPLSSE 554
+ P + P + + P SP +S PSP + P S PT P+ Y+P +S
Sbjct: 303 SPPSNSPTSTSPPPQPSPPNS-TSPSPPQTP-------TRKTSHSPTYPLPSPYSPATST 458
Query: 555 PIIHAQP 561
P
Sbjct: 459 SATEPYP 479
Score = 28.1 bits (61), Expect = 6.7
Identities = 19/84 (22%), Positives = 34/84 (39%), Gaps = 1/84 (1%)
Frame = +3
Query: 660 ILANPLQEADPLAQQAHPAPDQQEPIQPEPEPEQSVSNQSSVRSPH-PLVETSEPHLGTS 718
+LA+P + P+P P P P+ S N +S P P +TS
Sbjct: 249 VLASPSTSSTAPTTPPSPSPPSNSPTSTSPPPQPSPPNSTSPSPPQTPTRKTS------- 407
Query: 719 KPNAQTFNIGSPQGASEAHNSNHP 742
++ T+ + SP + + ++ P
Sbjct: 408 --HSPTYPLPSPYSPATSTSATEP 473
>TC17551 weakly similar to UP|AAR24662 (AAR24662) At5g26960, partial (22%)
Length = 505
Score = 35.4 bits (80), Expect = 0.042
Identities = 32/121 (26%), Positives = 50/121 (40%)
Frame = +3
Query: 497 PESQPAAQTTSPSHSPRSSFFQPSPTEAPLWNLLQNQPSRSEEPTSPITIQYNPLSSEPI 556
P + T+SP+ P S PSP+ A + P+ S PTSP + S+ P
Sbjct: 135 PSLTTSFSTSSPASHPLPSL--PSPSSAAVG------PTSSTPPTSPTSAAAATSSATPP 290
Query: 557 IHAQPEPTHTEPEPRTSDHSVPRASERLALKPTDTDSSTAFTPVSFPTNVADSSPSNNSE 616
+ P + + P P ++ + P +S P T S + + S PT + S S
Sbjct: 291 SPSPPPTSASPPPPSSTPNGTPPSS-----SPATTPSPSRPSTPSSPTRASLPSAPGFSS 455
Query: 617 S 617
S
Sbjct: 456 S 458
Score = 33.1 bits (74), Expect = 0.21
Identities = 24/85 (28%), Positives = 36/85 (42%)
Frame = +3
Query: 661 LANPLQEADPLAQQAHPAPDQQEPIQPEPEPEQSVSNQSSVRSPHPLVETSEPHLGTSKP 720
L +P+Q P P+P+ + + P P + S +S + HPL P P
Sbjct: 48 LVSPIQTTPP-PPPPPPSPNHRNNLPPPFPPSLTTSFSTSSPASHPLPSLPSPSSAAVGP 224
Query: 721 NAQTFNIGSPQGASEAHNSNHPASP 745
+ T SP A+ A +S P SP
Sbjct: 225 TSST-PPTSPTSAAAATSSATPPSP 296
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.313 0.129 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,415,750
Number of Sequences: 28460
Number of extensions: 259301
Number of successful extensions: 2290
Number of sequences better than 10.0: 218
Number of HSP's better than 10.0 without gapping: 1986
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2232
length of query: 939
length of database: 4,897,600
effective HSP length: 99
effective length of query: 840
effective length of database: 2,080,060
effective search space: 1747250400
effective search space used: 1747250400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0128.10


