

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0127b.3
(823 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
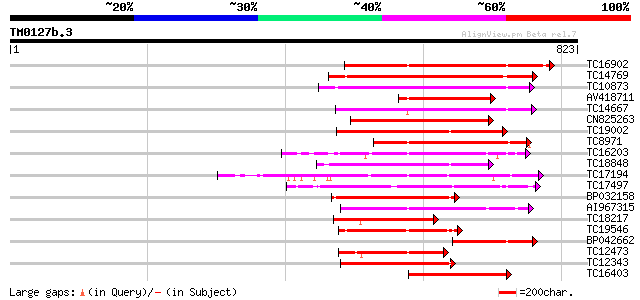
Score E
Sequences producing significant alignments: (bits) Value
TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-... 350 7e-97
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 261 2e-70
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 243 9e-65
AV418711 226 1e-59
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 206 2e-53
CN825263 202 2e-52
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 195 3e-50
TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P... 186 2e-47
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 182 2e-46
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 181 3e-46
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 173 9e-44
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 171 6e-43
BP032158 163 9e-41
AI967315 162 3e-40
TC18217 weakly similar to UP|Q9FLJ8 (Q9FLJ8) Receptor protein ki... 158 3e-39
TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine k... 155 2e-38
BP042662 149 2e-36
TC12473 similar to UP|Q9M068 (Q9M068) Protein kinase-like protei... 148 4e-36
TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine prote... 147 9e-36
TC16403 homologue to UP|Q9LKY4 (Q9LKY4) Pti1 kinase-like protein... 145 3e-35
>TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like
protein, partial (46%)
Length = 941
Score = 350 bits (897), Expect = 7e-97
Identities = 174/305 (57%), Positives = 226/305 (74%)
Frame = +3
Query: 487 EDLVIGSGGFGKVYRGVFKDETKVAVKRGSCQSQQGLAEFRTEIEMLSQFRHRHLVSLIG 546
E L++G GGFGKVY G TKVA+KRG+ S+QG+ EF+TEIEMLS+ RHRHLVSLIG
Sbjct: 3 EALLLGVGGFGKVYYGEVDGGTKVAIKRGNPLSEQGVHEFQTEIEMLSKLRHRHLVSLIG 182
Query: 547 YCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQRLEICIGAAKGLHYLHTGSNKA 606
YC E +E I++Y++M G+L++HL+ + L WKQRLEICIGAA+GLHYLHTG+
Sbjct: 183 YCEENTEMILVYDHMAYGTLREHLYKTQKPP--LPWKQRLEICIGAARGLHYLHTGAKYT 356
Query: 607 IIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYVSTAVKGSFGYLDPEYLITQQLT 666
IIHRDVK+ NILLDE +AKV+DFGLSKTGP +D +VST VKGSFGYLDPEY QQLT
Sbjct: 357 IIHRDVKTTNILLDEKWVAKVSDFGLSKTGPTLDNTHVSTVVKGSFGYLDPEYFRRQQLT 536
Query: 667 EKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIMRWQERSTIEELVDHHLPGSGQI 726
+KSDVYSFGVV+FE+LC RP ++PSL +E+++L EW + +++++D +L G+I
Sbjct: 537 DKSDVYSFGVVLFEILCARPALNPSLAKEQVSLAEWASHCYNKGILDQILDPYL--KGKI 710
Query: 727 KSESLHEFVDTAKKCLAEHGINRPSMGDVLWHLEYALRLEGVDERSNHGGDVSSQIHRSD 786
E +F +TA KC+++ GI RPSMGDVLW+LE+AL+L+ E S +G IH D
Sbjct: 711 APECFKKFAETAMKCVSDQGIERPSMGDVLWNLEFALQLQESAEESGNG---FGGIHGED 881
Query: 787 TGLSA 791
L A
Sbjct: 882 EPLFA 896
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 261 bits (668), Expect = 2e-70
Identities = 141/305 (46%), Positives = 190/305 (62%), Gaps = 1/305 (0%)
Frame = +3
Query: 463 FFTNSKIGYRLPLAVIQEATDNFSEDLVIGSGGFGKVYRGVFKDETKVAVKRGSCQSQQG 522
FF S L I+ AT+ + +IG GGFG VYRG D +VAVK S S QG
Sbjct: 2103 FFIKSVSIQAFTLEYIEVATERYKT--LIGEGGFGSVYRGTLNDGQEVAVKVRSSTSTQG 2276
Query: 523 LAEFRTEIEMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSW 582
EF E+ +LS +H +LV L+GYCNE ++I++Y +M GSL+D L+G A L W
Sbjct: 2277 TREFDNELNLLSAIQHENLVPLLGYCNESDQQILVYPFMSNGSLQDRLYGEPAKRKILDW 2456
Query: 583 KQRLEICIGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKR 642
RL I +GAA+GL YLHT +++IHRD+KS+NILLD ++ AKVADFG SK P
Sbjct: 2457 PTRLSIALGAARGLAYLHTFPGRSVIHRDIKSSNILLDHSMCAKVADFGFSKYAPQEGDS 2636
Query: 643 YVSTAVKGSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEW 702
YVS V+G+ GYLDPEY TQQL+EKSDV+SFGVV+ E++ GR ++ PR + +LVEW
Sbjct: 2637 YVSLEVRGTAGYLDPEYYKTQQLSEKSDVFSFGVVLLEIVSGREPLNIKRPRTEWSLVEW 2816
Query: 703 IMRWQERSTIEELVDHHLPG-SGQIKSESLHEFVDTAKKCLAEHGINRPSMGDVLWHLEY 761
+ S ++E+VD PG G +E++ V+ A +CL RPSM ++ LE
Sbjct: 2817 ATPYIRGSKVDEIVD---PGIKGGYHAEAMWRVVEVALQCLEPFSTYRPSMVAIVRELED 2987
Query: 762 ALRLE 766
AL +E
Sbjct: 2988 ALIIE 3002
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 243 bits (620), Expect = 9e-65
Identities = 135/314 (42%), Positives = 187/314 (58%), Gaps = 1/314 (0%)
Frame = +1
Query: 449 KGDHGATSNYDGTAFFTNSKIGYRLPLAVIQEATDNFSEDLVIGSGGFGKVYRGVFKDET 508
KG ++SN + G+R + +AT NF E +IG GGFGKVY+G
Sbjct: 355 KGKGVSSSNGSSNGKTAAASFGFR----ELADATRNFKEANLIGEGGFGKVYKGRLTTGE 522
Query: 509 KVAVKRGSCQSQQGLAEFRTEIEMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKD 568
VAVK+ S +QG EF E+ MLS H +LV LIGYC + +R+++YEYM GSL+D
Sbjct: 523 AVAVKQLSHDGRQGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLED 702
Query: 569 HLFGSNADATCLSWKQRLEICIGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVA 628
HLF + D L+W R+++ +GAA+GL YLH ++ +I+RD+KSANILLD K++
Sbjct: 703 HLFELSHDKEPLNWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLS 882
Query: 629 DFGLSKTGPDIDKRYVSTAVKGSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVI 688
DFGL+K GP D +VST V G++GY PEY ++ +LT KSD+YSFGVV+ E+L GR I
Sbjct: 883 DFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAI 1062
Query: 689 DPSLPREKMNLVEWIMRW-QERSTIEELVDHHLPGSGQIKSESLHEFVDTAKKCLAEHGI 747
D S + NLV W + +R +VD L G+ S LH+ + CL E
Sbjct: 1063DTSRRPGEQNLVSWARPYFSDRRRFGHMVDPLL--QGRFPSRCLHQAIAITAMCLQEQPK 1236
Query: 748 NRPSMGDVLWHLEY 761
RP + D++ LEY
Sbjct: 1237FRPLITDIVVALEY 1278
>AV418711
Length = 419
Score = 226 bits (576), Expect = 1e-59
Identities = 109/141 (77%), Positives = 125/141 (88%)
Frame = +3
Query: 565 SLKDHLFGSNADATCLSWKQRLEICIGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLM 624
+LK HL+GS + LSWK+RL+ICIG+A+GLHYLHTG KA+IHRDVKSANILLD+NLM
Sbjct: 3 TLKSHLYGSGFPS--LSWKERLDICIGSARGLHYLHTGYAKAVIHRDVKSANILLDDNLM 176
Query: 625 AKVADFGLSKTGPDIDKRYVSTAVKGSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCG 684
AKVADFGLSKTGP++D+ +VSTAVKGSFGYLDPEY QQLTEKSDVYSFGVV+FEVLC
Sbjct: 177 AKVADFGLSKTGPELDQTHVSTAVKGSFGYLDPEYFRRQQLTEKSDVYSFGVVLFEVLCA 356
Query: 685 RPVIDPSLPREKMNLVEWIMR 705
RPVIDPSLPRE +NL EW M+
Sbjct: 357 RPVIDPSLPREMVNLAEWAMK 419
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 206 bits (523), Expect = 2e-53
Identities = 117/298 (39%), Positives = 169/298 (56%), Gaps = 6/298 (2%)
Frame = +2
Query: 473 LPLAVIQEATDNFSEDLVIGSGGFGKVYRGVFKDETKVAVKRGSCQSQ-QGLAEFRTEIE 531
L L ++E TDNF +IG G +G+VY D VAVK+ S+ + EF T++
Sbjct: 323 LSLDELKEKTDNFGSKALIGEGSYGRVYYATLNDGNAVAVKKLDVSSEPETNNEFLTQVS 502
Query: 532 MLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNA-----DATCLSWKQRL 586
M+S+ ++ + V L GYC E + R++ YE+ GSL D L G L W QR+
Sbjct: 503 MVSRLKNDNFVELHGYCVEGNLRVLAYEFATMGSLHDILHGRKGVQGAQPGPTLDWIQRV 682
Query: 587 EICIGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYVST 646
I + AA+GL YLH AIIHRD++S+N+L+ E+ AK+ADF LS PD+ R ST
Sbjct: 683 RIAVDAARGLEYLHEKVQPAIIHRDIRSSNVLIFEDYKAKIADFNLSNQAPDMAARLHST 862
Query: 647 AVKGSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIMRW 706
V G+FGY PEY +T QLT+KSDVYSFGVV+ E+L GR +D ++PR + +LV W
Sbjct: 863 RVLGTFGYHAPEYAMTGQLTQKSDVYSFGVVLLELLTGRKPVDHTMPRGQQSLVTWATPR 1042
Query: 707 QERSTIEELVDHHLPGSGQIKSESLHEFVDTAKKCLAEHGINRPSMGDVLWHLEYALR 764
+++ VD L G+ + + + A C+ RP+M V+ L+ L+
Sbjct: 1043LSEDKVKQCVDPKL--KGEYPPKGVAKLAAVAALCVQYEAEFRPNMSIVVKALQPLLK 1210
>CN825263
Length = 663
Score = 202 bits (513), Expect = 2e-52
Identities = 102/208 (49%), Positives = 137/208 (65%)
Frame = +1
Query: 495 GFGKVYRGVFKDETKVAVKRGSCQSQQGLAEFRTEIEMLSQFRHRHLVSLIGYCNEQSER 554
GFG VY+G+ D VAVK Q+G EF E+EMLS+ HR+LV LIG C E+ R
Sbjct: 1 GFGLVYKGILNDGRDVAVKILKRDDQRGGREFLAEVEMLSRLHHRNLVKLIGICIEKQTR 180
Query: 555 IIIYEYMEKGSLKDHLFGSNADATCLSWKQRLEICIGAAKGLHYLHTGSNKAIIHRDVKS 614
+IYE + GS++ HL G++ + L W R++I +GAA+GL YLH SN +IHRD KS
Sbjct: 181 CLIYELVPNGSVESHLHGADKETGPLDWNARMKIALGAARGLAYLHEDSNPCVIHRDFKS 360
Query: 615 ANILLDENLMAKVADFGLSKTGPDIDKRYVSTAVKGSFGYLDPEYLITQQLTEKSDVYSF 674
+NILL+ + KV+DFGL++T D +++ST V G+FGYL PEY +T L KSDVYS+
Sbjct: 361 SNILLECDFTPKVSDFGLARTALDEGNKHISTHVMGTFGYLAPEYAMTGHLLVKSDVYSY 540
Query: 675 GVVMFEVLCGRPVIDPSLPREKMNLVEW 702
GVV+ E+L G +D S P + NLV W
Sbjct: 541 GVVLLELLTGTKPVDLSQPPGQENLVTW 624
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 195 bits (495), Expect = 3e-50
Identities = 100/248 (40%), Positives = 150/248 (60%)
Frame = +1
Query: 475 LAVIQEATDNFSEDLVIGSGGFGKVYRGVFKDETKVAVKRGSCQSQQGLAEFRTEIEMLS 534
L + AT+NF+ D +G GGFG VY G D +++AVKR S + EF E+E+L+
Sbjct: 34 LKELHSATNNFNYDNKLGEGGFGSVYWGQLWDGSQIAVKRLKVWSNKADMEFAVEVEILA 213
Query: 535 QFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQRLEICIGAAK 594
+ RH++L+SL GYC E ER+I+Y+YM SL HL G ++ L W +R+ I IG+A+
Sbjct: 214 RVRHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHLHGQHSSECLLDWNRRMNIAIGSAE 393
Query: 595 GLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYVSTAVKGSFGY 654
G+ YLH + IIHRD+K++N+LLD + A+VADFG +K PD +V+T VKG+ GY
Sbjct: 394 GIVYLHHQATPHIIHRDIKASNVLLDSDFQARVADFGFAKLIPD-GATHVTTRVKGTLGY 570
Query: 655 LDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIMRWQERSTIEE 714
L PEY + + E DV+SFG+++ E+ G+ ++ K ++ +W + E
Sbjct: 571 LAPEYAMLGKANECCDVFSFGILLLELASGKKPLEKLSSTVKRSINDWALPLACAKKFTE 750
Query: 715 LVDHHLPG 722
D L G
Sbjct: 751 FADPRLNG 774
>TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P14.15
{Arabidopsis thaliana;}, partial (37%)
Length = 1079
Score = 186 bits (471), Expect = 2e-47
Identities = 100/233 (42%), Positives = 152/233 (64%), Gaps = 4/233 (1%)
Frame = +3
Query: 529 EIEMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQRLEI 588
E+E+L++ HR+LV L+G+ ++ +ERI+I EY+ G+L++HL G L + QRLEI
Sbjct: 6 EVELLAKIDHRNLVKLLGFIDKGNERILITEYVPNGTLREHLDGLRGKI--LDFNQRLEI 179
Query: 589 CIGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGP-DIDKRYVSTA 647
I A GL YLH + K IIHRDVKS+NILL E++ AKVADFG ++ GP + D+ ++ST
Sbjct: 180 AIDVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFARLGPVNGDQTHISTK 359
Query: 648 VKGSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIMRWQ 707
VKG+ GYLDPEY+ T QLT KSDVYSFG+++ E+L GR ++ ++ + W R
Sbjct: 360 VKGTVGYLDPEYMKTHQLTPKSDVYSFGILLLEILTGRRPVELKKAADERVTLRWAFRKY 539
Query: 708 ERSTIEELVDHHLPGSGQIKSESLHEFVDTAKKCLAEHGINRPSM---GDVLW 757
++ EL+D + + + ++ L + +D + +C A +RP+M G+ LW
Sbjct: 540 NEGSVVELMDPLMEEA--VNADVLMKMLDLSFQCAAPIRTDRPNMKSVGEQLW 692
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 182 bits (461), Expect = 2e-46
Identities = 128/373 (34%), Positives = 202/373 (53%), Gaps = 11/373 (2%)
Frame = +2
Query: 395 SLVFDSEENGKTKVGLIVGLVAGSVVGFFAIVTAFVLLCRRRSRRRNSLHIGDEKGDHGA 454
S+++DS + K + +V G A+ TA +L+ ++H+ ++ H A
Sbjct: 1985 SVLYDSLRKTRAKTARVRAIVIG-----IALATAVLLVAV-------TVHVVRKRRLHRA 2128
Query: 455 TSNYDGTAFFTNSKIGYRLPLAVIQEATDNFSEDLVIGSGGFGKVYRGVFKDETKVAVKR 514
+ + TAF RL + ++ + E+ +IG GG G VYRG + T VA+KR
Sbjct: 2129 QA-WKLTAF-------QRLEIKA-EDVVECLKEENIIGKGGAGIVYRGSMPNGTDVAIKR 2281
Query: 515 ----GSCQSQQGLAEFRTEIEMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHL 570
GS ++ G FR EIE L + RHR+++ L+GY + + +++YEYM GSL + L
Sbjct: 2282 LVGQGSGRNDYG---FRAEIETLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWL 2452
Query: 571 FGSNADATCLSWKQRLEICIGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADF 630
G+ L W+ R +I + AA+GL Y+H + IIHRDVKS NILLD + A VADF
Sbjct: 2453 HGAKGGH--LRWEMRYKIAVEAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADF 2626
Query: 631 GLSKTGPDIDKRYVSTAVKGSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDP 690
GL+K D +++ GS+GY+ PEY T ++ EKSDVYSFGVV+ E++ GR +
Sbjct: 2627 GLAKFLYDPGASQSMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGE 2806
Query: 691 SLPREKMNLVEWIMRW-------QERSTIEELVDHHLPGSGQIKSESLHEFVDTAKKCLA 743
+ +++V W+ + + + + +VD L SG + +H F + A C+
Sbjct: 2807 F--GDGVDIVGWVNKTMSELSQPSDTALVLAVVDPRL--SGYPLTSVIHMF-NIAMMCVK 2971
Query: 744 EHGINRPSMGDVL 756
E G RP+M +V+
Sbjct: 2972 EMGPARPTMREVV 3010
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 181 bits (460), Expect = 3e-46
Identities = 99/258 (38%), Positives = 152/258 (58%), Gaps = 1/258 (0%)
Frame = +2
Query: 446 GDEKGDHGATSNYDGTAFFTNSKIGYRL-PLAVIQEATDNFSEDLVIGSGGFGKVYRGVF 504
G EK + G TS F + +R+ + AT FS+D +G GGFG VY G
Sbjct: 203 GSEKVEEGPTS-------FGSVNNSWRIFTYKELHAATGGFSDDNKLGEGGFGSVYWGRT 361
Query: 505 KDETKVAVKRGSCQSQQGLAEFRTEIEMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKG 564
D ++AVK+ + + EF E+E+L + RH++L+ L GYC +R+I+Y+YM
Sbjct: 362 SDGLQIAVKKLKAMNSKAEMEFAVEVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNL 541
Query: 565 SLKDHLFGSNADATCLSWKQRLEICIGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLM 624
SL HL G A L+W++R++I IG+A+G+ YLH IIHRD+K++N+LL+ +
Sbjct: 542 SLLSHLHGQFAVEVQLNWQKRMKIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLNSDFE 721
Query: 625 AKVADFGLSKTGPDIDKRYVSTAVKGSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCG 684
VADFG +K P+ +++T VKG+ GYL PEY + +++E DVYSFG+++ E++ G
Sbjct: 722 PLVADFGFAKLIPE-GVSHMTTRVKGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTG 898
Query: 685 RPVIDPSLPREKMNLVEW 702
R I+ K + EW
Sbjct: 899 RKPIEKLPGGVKRTITEW 952
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 173 bits (439), Expect = 9e-44
Identities = 153/520 (29%), Positives = 246/520 (46%), Gaps = 47/520 (9%)
Frame = +1
Query: 302 DYFVRFHFCDIVNLPPNTLFNVYLNSWLVSTVDLGKETSDKAGVPYYMDAVTRTSGASHG 361
D RF+ D N+P N NV +N G K +G
Sbjct: 493 DLLKRFNSYDPKNIPVNAKVNVTVNC------SCGNSQVSK----------------DYG 606
Query: 362 LNVSVGTYSISEASSPEAILN--GLEIMKISNSKDSLVFDSEEN-----GKTKVGLIV-- 412
L + TY I + + I N L+ I + S+ F + G+ K G+ V
Sbjct: 607 LFI---TYPIRPGDTLQDIANQSSLDAGLIQSFNPSVNFSKDSGIAFIPGRYKNGVYVPL 777
Query: 413 -----GLVAGSVVG------FFAIVTAFVLLCRRRSRRRN--------SLHIGDEKGDHG 453
GL +G+ VG F ++ AF + R + + S+ + + G+
Sbjct: 778 YHRTAGLASGAAVGISIAGTFVLLLLAFCMYVRYQKKEEEKAKLPTDISMALSTQDGNAS 957
Query: 454 ATSNYD-------GTAF---FTNSKIGYRLPLAV--IQEATDNFSEDLVIGSGGFGKVYR 501
+++ Y+ GTA T+ + + + + +AT+NFS D IG GGFG VY
Sbjct: 958 SSAEYETSGSSGPGTASATGLTSIMVAKSMEFSYQELAKATNNFSLDNKIGQGGFGAVYY 1137
Query: 502 GVFKDETKVAVKRGSCQSQQGLAEFRTEIEMLSQFRHRHLVSLIGYCNEQSERIIIYEYM 561
+ + K A+K+ Q+ EF E+++L+ H +LV LIGYC E S ++YE++
Sbjct: 1138AELRGK-KTAIKKMDVQAS---TEFLCELKVLTHVHHLNLVRLIGYCVEGS-LFLVYEHI 1302
Query: 562 EKGSLKDHLFGSNADATCLSWKQRLEICIGAAKGLHYLHTGSNKAIIHRDVKSANILLDE 621
+ G+L +L GS + L W R++I + AA+GL Y+H + IHRDVKSANIL+D+
Sbjct: 1303DNGNLGQYLHGSGKEP--LPWSSRVQIALDAARGLEYIHEHTVPVYIHRDVKSANILIDK 1476
Query: 622 NLMAKVADFGLSKTGPDIDKRYVSTAVKGSFGYLDPEYLITQQLTEKSDVYSFGVVMFEV 681
NL KVADFGL+K ++ + T + G+FGY+ PEY ++ K DVY+FGVV+FE+
Sbjct: 1477NLRGKVADFGLTKL-IEVGNSTLQTRLVGTFGYMPPEYAQYGDISPKIDVYAFGVVLFEL 1653
Query: 682 LCGRPVI--DPSLPREKMNLV----EWIMRWQERSTIEELVDHHLPGSGQIKSESLHEFV 735
+ + + L E LV E + + + +LVD L + I +S+ +
Sbjct: 1654ISAKNAVLKTGELVAESKGLVALFEEALNKSDPCDALRKLVDPRLGENYPI--DSVLKIA 1827
Query: 736 DTAKKCLAEHGINRPSMGDVLWHLEYALRL-EGVDERSNH 774
+ C ++ + RPSM ++ L L E D+ S++
Sbjct: 1828QLGRACTRDNPLLRPSMRSLVVALMTLSSLTEDCDDESSY 1947
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 171 bits (432), Expect = 6e-43
Identities = 118/371 (31%), Positives = 190/371 (50%), Gaps = 3/371 (0%)
Frame = +1
Query: 403 NGKTKVGLIVGLVAGSVVGFFAIVTAFVLLCRRRSRRRNSLHIGDEKGDHGATSNYDGTA 462
N K G +V ++A V F I+ + C +R + + GDE G+ G +++
Sbjct: 1447 NTKKLAGSLVVIIA--FVIFITILGLAISTCIQRKKNKR----GDE-GEIGIINHWKDKR 1605
Query: 463 FFTNSKIGYRLPLAVIQEATDNFSEDLVIGSGGFGKVYRGVFKDETKVAVKRGSCQSQQG 522
+ + + I AT++FS +G GGFG VY+G+ + ++AVKR S S QG
Sbjct: 1606 GDEDIDLATIFDFSTISSATNHFSLSNKLGEGGFGPVYKGLLANGQEIAVKRLSNTSGQG 1785
Query: 523 LAEFRTEIEMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSW 582
+ EF+ EI+++++ +HR+LV L G Q E + K + + + + W
Sbjct: 1786 MEEFKNEIKLIARLQHRNLVKLFGCSVHQDEN-------SHANKKMKILLDSTRSKLVDW 1944
Query: 583 KQRLEICIGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSK--TGPDID 640
+RL+I G A+GL YLH S IIHRD+K++NILLD+ + K++DFGL++ G ++
Sbjct: 1945 NKRLQIIDGIARGLLYLHQDSRLRIIHRDLKTSNILLDDEMNPKISDFGLARIFIGDQVE 2124
Query: 641 KRYVSTAVKGSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLV 700
R + V G++GY+ PEY + + KSDV+SFGV++ E++ G+ + P +NL+
Sbjct: 2125 AR--TKRVMGTYGYMPPEYAVHGSFSIKSDVFSFGVIVLEIISGKKIGRFYDPHHHLNLL 2298
Query: 701 EWIMR-WQERSTIEELVDHHLPGSGQIKSESLHEFVDTAKKCLAEHGINRPSMGDVLWHL 759
R W E + ELVD L + + ++ A C+ NRP M L
Sbjct: 2299 SHAWRLWIEERPL-ELVDELL--DDPVIPTEILRYIHVALLCVQRRPENRPDM------L 2451
Query: 760 EYALRLEGVDE 770
L L G E
Sbjct: 2452 SIVLMLNGEKE 2484
>BP032158
Length = 555
Score = 163 bits (413), Expect = 9e-41
Identities = 84/186 (45%), Positives = 121/186 (64%)
Frame = +2
Query: 468 KIGYRLPLAVIQEATDNFSEDLVIGSGGFGKVYRGVFKDETKVAVKRGSCQSQQGLAEFR 527
K GY L I+ AT+NF IG GGFG VY+GV + +AVK+ S +S+QG EF
Sbjct: 2 KTGY-FSLRQIKAATNNFDPANKIGEGGFGPVYKGVLSEGDVIAVKQLSSKSKQGNREFI 178
Query: 528 TEIEMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQRLE 587
EI M+S +H +LV L G C E ++ +++YEYME SL LFG+ L+W+ R++
Sbjct: 179 NEIGMISALQHPNLVKLYGCCIEGNQLLLVYEYMENNSLARALFGNEEQKLNLNWRTRMK 358
Query: 588 ICIGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYVSTA 647
IC+G AKGL YLH S I+HRD+K+ N+LLD++L AK++DFGL+K + + ++ST
Sbjct: 359 ICVGIAKGLAYLHEESRLKIVHRDIKATNVLLDKDLKAKISDFGLAKLDEE-ENTHISTR 535
Query: 648 VKGSFG 653
+ G+ G
Sbjct: 536 IAGTIG 553
>AI967315
Length = 1308
Score = 162 bits (409), Expect = 3e-40
Identities = 96/282 (34%), Positives = 155/282 (54%), Gaps = 2/282 (0%)
Frame = +1
Query: 481 ATDNFSEDLVIGSGGFGKVYRGVFKDETKVAVKR--GSCQSQQGLAEFRTEIEMLSQFRH 538
AT+ FS + ++G GG+ +VY+G + ++AVKR +C+ ++ EF TEI + H
Sbjct: 136 ATNGFSSENMVGKGGYAEVYKGRLESGDEIAVKRLTRTCRDERKEKEFLTEIGTIGHVCH 315
Query: 539 RHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQRLEICIGAAKGLHY 598
+++ L+G C + +++E GS+ + L WK R +I +G A+GLHY
Sbjct: 316 SNVMPLLGCCIDNG-LYLVFELSTVGSVASLIHDEKMAP--LDWKTRYKIVLGTARGLHY 486
Query: 599 LHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYVSTAVKGSFGYLDPE 658
LH G + IIHRD+K++NILL E+ +++DFGL+K P + ++G+FG+L PE
Sbjct: 487 LHKGCQRRIIHRDIKASNILLTEDFEPQISDFGLAKWLPSQWTHHSIAPIEGTFGHLAPE 666
Query: 659 YLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEWIMRWQERSTIEELVDH 718
Y + + EK+DV++FGV + EV+ GR +D S +L W + IE+LVD
Sbjct: 667 YYMHGVVDEKTDVFAFGVFLLEVISGRKPVDGS----HQSLHTWAKPILSKWEIEKLVDP 834
Query: 719 HLPGSGQIKSESLHEFVDTAKKCLAEHGINRPSMGDVLWHLE 760
L G + + F A C+ RP+M +VL +E
Sbjct: 835 RLEGCYDVTQFNRVAF--AASLCIRASSTWRPTMSEVLEVME 954
>TC18217 weakly similar to UP|Q9FLJ8 (Q9FLJ8) Receptor protein kinase-like
protein, partial (15%)
Length = 688
Score = 158 bits (400), Expect = 3e-39
Identities = 81/156 (51%), Positives = 108/156 (68%), Gaps = 4/156 (2%)
Frame = +3
Query: 471 YRLPLAVIQEATDNFSEDLVIGSGGFGKVYRGVFKDE----TKVAVKRGSCQSQQGLAEF 526
+ LA +++AT+NF E+ VIG G GKVY+G + + T VAVK+ S GL EF
Sbjct: 219 HHFSLADLKKATNNFDENNVIGDRGNGKVYKGFLQLDHVATTVVAVKQMDNSSFWGLKEF 398
Query: 527 RTEIEMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQRL 586
+ EIE+L Q RH +L++LIG+C + E+II++EYM GSL D L S LSWK+R+
Sbjct: 399 KNEIELLCQLRHPNLITLIGFCLHKDEKIIVFEYMSNGSLADRLLNSRDARKPLSWKKRI 578
Query: 587 EICIGAAKGLHYLHTGSNKAIIHRDVKSANILLDEN 622
EICIGAA+GLHYLH+G+ + I HR +K ANILLDEN
Sbjct: 579 EICIGAARGLHYLHSGAKRNIFHRGIKPANILLDEN 686
>TC19546 similar to UP|Q40543 (Q40543) Protein-serine/threonine kinase ,
partial (46%)
Length = 731
Score = 155 bits (392), Expect = 2e-38
Identities = 80/180 (44%), Positives = 122/180 (67%)
Frame = +1
Query: 478 IQEATDNFSEDLVIGSGGFGKVYRGVFKDETKVAVKRGSCQSQQGLAEFRTEIEMLSQFR 537
IQ+AT NF+ ++G G FG VY+ VAVK + S+QG EF+TE+ +L +
Sbjct: 214 IQKATQNFTT--ILGQGSFGTVYKATMPTGEVVAVKVLAPNSKQGEHEFQTEVHLLGRLH 387
Query: 538 HRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQRLEICIGAAKGLH 597
HR+LV+L+G+C ++ +RI++Y++M GSL + L+G + LSW +RL+I + + G+
Sbjct: 388 HRNLVNLVGFCVDKGQRILVYQFMSNGSLANLLYGEEKE---LSWDERLQIAMDISHGIE 558
Query: 598 YLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYVSTAVKGSFGYLDP 657
YLH G+ +IHRD+KSANILLD+++ A VADFGLSK D R ++ +KG++GY+DP
Sbjct: 559 YLHEGAVPPVIHRDLKSANILLDDSMRAMVADFGLSKE-EIFDGR--NSGLKGTYGYMDP 729
>BP042662
Length = 484
Score = 149 bits (375), Expect = 2e-36
Identities = 74/123 (60%), Positives = 91/123 (73%)
Frame = -3
Query: 643 YVSTAVKGSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNLVEW 702
YV T VKG+FGYLDPEY +QQLTEKSDVYSFGVV+ EVLC RP I+ S PRE++NL EW
Sbjct: 473 YVITDVKGTFGYLDPEYFRSQQLTEKSDVYSFGVVLLEVLCARPAIESSFPREQVNLAEW 294
Query: 703 IMRWQERSTIEELVDHHLPGSGQIKSESLHEFVDTAKKCLAEHGINRPSMGDVLWHLEYA 762
M + + + E+VD + GQI SL +F DT +KCL E+G +RP+M DVLW LEYA
Sbjct: 293 GMLCKNKGMLGEIVDPAI--KGQIDQNSLKKFSDTVEKCLQENGCDRPTMRDVLWDLEYA 120
Query: 763 LRL 765
L+L
Sbjct: 119 LQL 111
>TC12473 similar to UP|Q9M068 (Q9M068) Protein kinase-like protein, partial
(42%)
Length = 946
Score = 148 bits (373), Expect = 4e-36
Identities = 79/171 (46%), Positives = 115/171 (67%), Gaps = 11/171 (6%)
Frame = +3
Query: 478 IQEATDNFSEDLVIGSGGFGKVYRGVFKDETK-----------VAVKRGSCQSQQGLAEF 526
++ AT +F D +IG GGFGKVY+G + DE K VAVK+ + +S QG E+
Sbjct: 435 LKSATKSFKADALIGEGGFGKVYKG-WLDEKKLSPTKPGSGIMVAVKKLNPESMQGFHEW 611
Query: 527 RTEIEMLSQFRHRHLVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQRL 586
++EI L + H +LV L+GYC + E +++YE+M +GSL++HLF N ++ LSW RL
Sbjct: 612 QSEINFLGRISHPNLVKLLGYCRDDEEFLLVYEFMPRGSLENHLFRRNTES--LSWNTRL 785
Query: 587 EICIGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGP 637
+I IGAA+GL +LH+ K +I+RD K++NILLD N AK++ FGL+K GP
Sbjct: 786 KIAIGAARGLAFLHS-LXKIVIYRDFKASNILLDGNYNAKISXFGLTKFGP 935
>TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine protein kinase
Cdk9 (Cyclin-dependent kinase Cdk9) , partial (7%)
Length = 723
Score = 147 bits (370), Expect = 9e-36
Identities = 76/166 (45%), Positives = 111/166 (66%)
Frame = +3
Query: 481 ATDNFSEDLVIGSGGFGKVYRGVFKDETKVAVKRGSCQSQQGLAEFRTEIEMLSQFRHRH 540
AT NF +G GGFG V++G D ++AVK+ S +S QG +F E ++L++ +HR+
Sbjct: 231 ATKNFHAVNKLGEGGFGPVFKGKLNDGREIAVKKLSRRSNQGRTQFINEAKLLTRVQHRN 410
Query: 541 LVSLIGYCNEQSERIIIYEYMEKGSLKDHLFGSNADATCLSWKQRLEICIGAAKGLHYLH 600
+VSL GYC SE++++YEY+ + SL LF S L WK+R +I G A+GL YLH
Sbjct: 411 VVSLFGYCAHGSEKLLVYEYVPRESLDKLLFRSQKKEQ-LDWKRRFDIISGVARGLLYLH 587
Query: 601 TGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDIDKRYVST 646
S+ IIHRD+K+ANILLDE + K+ADFGL++ P+ D+ +V+T
Sbjct: 588 EDSHDCIIHRDIKAANILLDEKWVPKIADFGLARIFPE-DQTHVNT 722
>TC16403 homologue to UP|Q9LKY4 (Q9LKY4) Pti1 kinase-like protein, partial
(55%)
Length = 876
Score = 145 bits (365), Expect = 3e-35
Identities = 75/149 (50%), Positives = 98/149 (65%)
Frame = +3
Query: 580 LSWKQRLEICIGAAKGLHYLHTGSNKAIIHRDVKSANILLDENLMAKVADFGLSKTGPDI 639
LSW QR++I +GAA+GL YLH + IIHR +KS+NILL ++ +AK+ADF LS PD
Sbjct: 36 LSWTQRVKIAVGAARGLEYLHEKAETHIIHRYIKSSNILLFDDDVAKIADFDLSNQAPDA 215
Query: 640 DKRYVSTAVKGSFGYLDPEYLITQQLTEKSDVYSFGVVMFEVLCGRPVIDPSLPREKMNL 699
R ST V G+FGY PEY +T QLT KSDVYSFGVV+ E+L GR +D +LPR + +L
Sbjct: 216 AARLHSTRVLGTFGYHAPEYAMTGQLTSKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSL 395
Query: 700 VEWIMRWQERSTIEELVDHHLPGSGQIKS 728
V W +++ VD L G +KS
Sbjct: 396 VTWATPKLSEDKVKQCVDARLKGEYPVKS 482
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.135 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,558,040
Number of Sequences: 28460
Number of extensions: 187330
Number of successful extensions: 1437
Number of sequences better than 10.0: 301
Number of HSP's better than 10.0 without gapping: 1274
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1288
length of query: 823
length of database: 4,897,600
effective HSP length: 98
effective length of query: 725
effective length of database: 2,108,520
effective search space: 1528677000
effective search space used: 1528677000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0127b.3


