

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
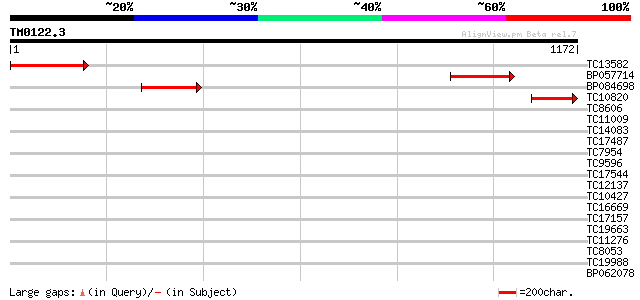
Query= TM0122.3
(1172 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC13582 similar to UP|Q9M9Y0 (Q9M9Y0) F4H5.19 protein, partial (9%) 320 1e-87
BP057714 259 1e-69
BP084698 255 3e-68
TC10820 weakly similar to UP|Q9M9Y0 (Q9M9Y0) F4H5.19 protein, pa... 184 7e-47
TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%) 40 0.002
TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Ar... 35 0.090
TC14083 similar to UP|CRTC_RICCO (P93508) Calreticulin precursor... 34 0.15
TC17487 UP|CAE45597 (CAE45597) SAR DNA-binding protein-like prot... 33 0.20
TC7954 similar to UP|Q7XAU4 (Q7XAU4) Ethylene responsive protein... 33 0.20
TC9596 similar to UP|AAR24722 (AAR24722) At5g49400, partial (66%) 33 0.20
TC17544 similar to PIR|T04919|T04919 DNA-binding protein homolog... 33 0.26
TC12137 similar to UP|Q8L7W3 (Q8L7W3) At2g29210/F16P2.41, partia... 33 0.26
TC10427 similar to UP|Q9AU06 (Q9AU06) NADPH-cytochrome P450 oxyd... 32 0.45
TC16669 similar to UP|Q9SLT9 (Q9SLT9) ZCF37 protein (T30E16.15),... 32 0.59
TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly pr... 32 0.59
TC19663 similar to UP|Q8H6S8 (Q8H6S8) Translation initiation fac... 32 0.77
TC11276 weakly similar to UP|HDA1_CHICK (P56517) Histone deacety... 32 0.77
TC8053 similar to UP|Q94A20 (Q94A20) AT4g15470/dl3775w, partial ... 32 0.77
TC19988 similar to UP|CRT3_ARATH (O04153) Calreticulin 3 precurs... 31 1.3
BP062078 28 8.5
>TC13582 similar to UP|Q9M9Y0 (Q9M9Y0) F4H5.19 protein, partial (9%)
Length = 559
Score = 320 bits (819), Expect = 1e-87
Identities = 160/162 (98%), Positives = 160/162 (98%)
Frame = +1
Query: 1 MAVNDADQSNKAHRIRQSGAKSFKKKQNKKKKQDDDDGLIDKPQNPRAFAFSSTNKAKRL 60
MAVNDADQSNKAHRIRQSGAKSFKKKQNKKKKQDDDDGLIDKPQNPRAFAFSSTNKAKRL
Sbjct: 73 MAVNDADQSNKAHRIRQSGAKSFKKKQNKKKKQDDDDGLIDKPQNPRAFAFSSTNKAKRL 252
Query: 61 KSRAVEKEQRRLHVPITDRSYGEPAPYVIVVQGPPQVGKSLLIKSLVKHYTKHNLPEVRG 120
KSRAVEKEQRRLHVPITDRSYGEPAPYVIVVQGPPQVGKSL IKSLVKHYTKHNLPEVRG
Sbjct: 253 KSRAVEKEQRRLHVPITDRSYGEPAPYVIVVQGPPQVGKSLSIKSLVKHYTKHNLPEVRG 432
Query: 121 PITIVSGKQRRLQFVECPNDINGMIDAAKFADLALLLIDGSY 162
PITIVSGKQRRLQ VECPNDINGMIDAAKFADLALLLIDGSY
Sbjct: 433 PITIVSGKQRRLQLVECPNDINGMIDAAKFADLALLLIDGSY 558
>BP057714
Length = 402
Score = 259 bits (663), Expect = 1e-69
Identities = 129/133 (96%), Positives = 129/133 (96%)
Frame = -3
Query: 911 KKIKLVGYPCKIFKKTALIKEMFTSDLEIARFEGAAIRTVSGIRGQVKKAAKEEIGNQPK 970
KK GYPCKIFKKTALIKEMFTSDLEIARFEGAAIRTVSGIRGQVKKAAKEEIGNQPK
Sbjct: 400 KK*NWSGYPCKIFKKTALIKEMFTSDLEIARFEGAAIRTVSGIRGQVKKAAKEEIGNQPK 221
Query: 971 RKGGITKEGIARCTFEDKILMSDIVFLRAWTQVEVPQFYNPLTTALQPRDQTWKGMRTVA 1030
RKGGITKEGIARCTFEDKILMSDIVFLRAWTQVEVPQFYNPLTTALQPRDQTWKGMRTVA
Sbjct: 220 RKGGITKEGIARCTFEDKILMSDIVFLRAWTQVEVPQFYNPLTTALQPRDQTWKGMRTVA 41
Query: 1031 ELRRDHDLPVPVN 1043
ELRRDHDLPVPVN
Sbjct: 40 ELRRDHDLPVPVN 2
>BP084698
Length = 371
Score = 255 bits (651), Expect = 3e-68
Identities = 123/123 (100%), Positives = 123/123 (100%)
Frame = +3
Query: 273 KVHSNNKCDRKITLYGYLRGCNLKKGNKVHIAGVGDYSLAGVTALPDPCPLPSAAKKKGL 332
KVHSNNKCDRKITLYGYLRGCNLKKGNKVHIAGVGDYSLAGVTALPDPCPLPSAAKKKGL
Sbjct: 3 KVHSNNKCDRKITLYGYLRGCNLKKGNKVHIAGVGDYSLAGVTALPDPCPLPSAAKKKGL 182
Query: 333 RDKEKLFYAPMSGLGDLLYDKDAVYININDHFVQFTKVDDENSDITRKGNERDIGEVLVR 392
RDKEKLFYAPMSGLGDLLYDKDAVYININDHFVQFTKVDDENSDITRKGNERDIGEVLVR
Sbjct: 183 RDKEKLFYAPMSGLGDLLYDKDAVYININDHFVQFTKVDDENSDITRKGNERDIGEVLVR 362
Query: 393 SLQ 395
SLQ
Sbjct: 363 SLQ 371
>TC10820 weakly similar to UP|Q9M9Y0 (Q9M9Y0) F4H5.19 protein, partial (7%)
Length = 619
Score = 184 bits (467), Expect = 7e-47
Identities = 93/94 (98%), Positives = 94/94 (99%)
Frame = +2
Query: 1079 ETPKRNQPLLEQRRQKGVVMEPRERKVLALVQHIQLINHDKMKKRKLKENEKRKAHEAEK 1138
ETPKRNQPLLEQRRQKGVVMEPRERKVLALVQHIQLINHD+MKKRKLKENEKRKAHEAEK
Sbjct: 2 ETPKRNQPLLEQRRQKGVVMEPRERKVLALVQHIQLINHDEMKKRKLKENEKRKAHEAEK 181
Query: 1139 AKEEQLSKKRRREERREKYRTQDKSSKKMRRSEA 1172
AKEEQLSKKRRREERREKYRTQDKSSKKMRRSEA
Sbjct: 182 AKEEQLSKKRRREERREKYRTQDKSSKKMRRSEA 283
>TC8606 similar to UP|AAQ82688 (AAQ82688) Epa4p, partial (7%)
Length = 1008
Score = 40.4 bits (93), Expect = 0.002
Identities = 34/140 (24%), Positives = 56/140 (39%), Gaps = 7/140 (5%)
Frame = -2
Query: 409 MINLFAQSSEALAEAQGANMDVEQDGIIETVNYNEMDSDGGSESSDQDE-ADAMTGSGLP 467
+IN+ E ++++ D E + DG + D+D+ D G G
Sbjct: 686 LINMATTKGETKSQSEDKQHDAEGE-------------DGNDDEGDEDDDGDGPFGEGEE 546
Query: 468 DQAEDDAAG--DKSTNKDHLEEHIEFHNGRRRRRAIFGNDADQSDLMDSSGN----EDGS 521
D + +D AG + S NK + ++ E G G D + D D + ED
Sbjct: 545 DLSSEDGAGYGNNSNNKSNSKKAPEGGPGAGAGPEENGEDEEDEDGEDQEDDDDDDEDDD 366
Query: 522 ASDDGASSDSESLNEEEEDD 541
DDG + E EEE+++
Sbjct: 365 EEDDGGEDEEEEGVEEEDNE 306
>TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;}, partial (44%)
Length = 700
Score = 34.7 bits (78), Expect = 0.090
Identities = 39/170 (22%), Positives = 56/170 (32%)
Frame = +2
Query: 445 DSDGGSESSDQDEADAMTGSGLPDQAEDDAAGDKSTNKDHLEEHIEFHNGRRRRRAIFGN 504
D DG + D+D+ D G G D E+D EE GR
Sbjct: 38 DDDGEDDDGDEDDDDDAPGGGDDDDDEEDE-----------EEEGGVEGGR--------- 157
Query: 505 DADQSDLMDSSGNEDGSASDDGASSDSESLNEEEEDDVMGNVSKWKESLAERTLSQKTPS 564
G D DD D E EEEE++ +G E L RT++
Sbjct: 158 ----------GGGGDPDDDDDDDDDDDE---EEEEEEDLGT-----EYLVRRTVAA---- 271
Query: 565 LMQLVYGESTVNLISINKENDSSEDEESDGDFFEPIEEVKKKNVRDGLND 614
E E D +D ++D + +K++ +DG D
Sbjct: 272 ------AEDEEASSDFEPEEDEGDDNDNDDGEKAGVPSKRKRSDKDGSGD 403
Score = 33.5 bits (75), Expect = 0.20
Identities = 30/126 (23%), Positives = 47/126 (36%), Gaps = 9/126 (7%)
Frame = +2
Query: 429 DVEQDGIIETVNYNEMDSDGGSESSDQDEAD---------AMTGSGLPDQAEDDAAGDKS 479
D + DG + + ++ D G D DE D G G PD +DD D
Sbjct: 32 DDDDDGEDDDGDEDDDDDAPGGGDDDDDEEDEEEEGGVEGGRGGGGDPDDDDDD--DDDD 205
Query: 480 TNKDHLEEHIEFHNGRRRRRAIFGNDADQSDLMDSSGNEDGSASDDGASSDSESLNEEEE 539
++ EE + RR A ++ SD D + +DDG + S + +
Sbjct: 206 DEEEEEEEDLGTEYLVRRTVAAAEDEEASSDFEPEEDEGDDNDNDDGEKAGVPSKRKRSD 385
Query: 540 DDVMGN 545
D G+
Sbjct: 386 KDGSGD 403
>TC14083 similar to UP|CRTC_RICCO (P93508) Calreticulin precursor, partial
(92%)
Length = 1693
Score = 33.9 bits (76), Expect = 0.15
Identities = 31/110 (28%), Positives = 44/110 (39%), Gaps = 1/110 (0%)
Frame = +1
Query: 612 LNDGNVNTEDYSKCAQFMDKRWDKKDNEEIRNRFVTGNLAKAALRNGLPKATTEEENDD- 670
L D + T+D Q ++ W K+ + E KAA K EE +D
Sbjct: 1096 LFDNVLITDDPEYAKQVAEETWGKQKDAE-----------KAAFEEAEKKREEEESKEDP 1242
Query: 671 VYGDFEDLETGEKIENNQTDDATHNGDDLEAETRRLKKLALQEDGGNENE 720
D ED E TD+A+H+ DD E++T ED + NE
Sbjct: 1243 ADSDAEDEE--------DTDEASHDSDDAESKTE------AGEDSADANE 1350
Score = 30.0 bits (66), Expect = 2.2
Identities = 17/53 (32%), Positives = 25/53 (47%)
Frame = +1
Query: 1114 LINHDKMKKRKLKENEKRKAHEAEKAKEEQLSKKRRREERREKYRTQDKSSKK 1166
LI D +++ E K +AEKA E+ KKR EE +E D ++
Sbjct: 1111 LITDDPEYAKQVAEETWGKQKDAEKAAFEEAEKKREEEESKEDPADSDAEDEE 1269
>TC17487 UP|CAE45597 (CAE45597) SAR DNA-binding protein-like protein, complete
Length = 1794
Score = 33.5 bits (75), Expect = 0.20
Identities = 22/91 (24%), Positives = 47/91 (51%), Gaps = 3/91 (3%)
Frame = +1
Query: 1068 QERLPFASKPKETPKRNQPLLEQR---RQKGVVMEPRERKVLALVQHIQLINHDKMKKRK 1124
+E+ K ++ +++ PLL +K VV + +++K ++++L N D ++K
Sbjct: 1528 KEKKEKKKKKEKNEEKDVPLLADEDGDEEKEVVKKEKKKKRKESTENVELQNGDDAGEKK 1707
Query: 1125 LKENEKRKAHEAEKAKEEQLSKKRRREERRE 1155
+KRK H + E SKK+ ++++ E
Sbjct: 1708 ----KKRKKHAEPEESAEMPSKKKGKKKKSE 1788
>TC7954 similar to UP|Q7XAU4 (Q7XAU4) Ethylene responsive protein, partial
(75%)
Length = 1708
Score = 33.5 bits (75), Expect = 0.20
Identities = 16/51 (31%), Positives = 29/51 (56%)
Frame = +2
Query: 17 QSGAKSFKKKQNKKKKQDDDDGLIDKPQNPRAFAFSSTNKAKRLKSRAVEK 67
++G + FK + +++++DDD D + FAFS+T AK +S + K
Sbjct: 296 EAGFREFKDDSDFEEEEEDDDVAQDLLAGAKGFAFSATAAAKTKRSNSFSK 448
>TC9596 similar to UP|AAR24722 (AAR24722) At5g49400, partial (66%)
Length = 1196
Score = 33.5 bits (75), Expect = 0.20
Identities = 30/102 (29%), Positives = 42/102 (40%), Gaps = 5/102 (4%)
Frame = +2
Query: 445 DSDGGSESSD-----QDEADAMTGSGLPDQAEDDAAGDKSTNKDHLEEHIEFHNGRRRRR 499
DSD GS+S D + + ++TGS + D S++ D EE RRRRR
Sbjct: 644 DSDSGSDSEDSVFETESGSSSVTGSDYSSGSSSD-----SSSSDSEEE-------RRRRR 787
Query: 500 AIFGNDADQSDLMDSSGNEDGSASDDGASSDSESLNEEEEDD 541
+ G S SSDSES ++ + DD
Sbjct: 788 K-------KKTQKKKKGRRHKKYSSSSESSDSESASDSDSDD 892
Score = 29.3 bits (64), Expect = 3.8
Identities = 22/74 (29%), Positives = 35/74 (46%), Gaps = 2/74 (2%)
Frame = +2
Query: 468 DQAEDDAAGDKSTNKDHLEEHIEFHNGRRRRRAIFGNDADQSDLMDSSGNEDGSASD--D 525
+ +D KS++K +H R + G+D++ S SG+ + SD
Sbjct: 575 NDVDDKEEKAKSSSKKAKRKH--------RSDSDSGSDSEDSVFETESGSSSVTGSDYSS 730
Query: 526 GASSDSESLNEEEE 539
G+SSDS S + EEE
Sbjct: 731 GSSSDSSSSDSEEE 772
>TC17544 similar to PIR|T04919|T04919 DNA-binding protein homolog T9A21.10 -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(14%)
Length = 578
Score = 33.1 bits (74), Expect = 0.26
Identities = 23/75 (30%), Positives = 38/75 (50%)
Frame = +2
Query: 1082 KRNQPLLEQRRQKGVVMEPRERKVLALVQHIQLINHDKMKKRKLKENEKRKAHEAEKAKE 1141
K + L ++ Q VV+ + ++L L H+ L+ H M + K + KRK K KE
Sbjct: 284 KEIRSLQKEMLQVVVVVVVKPLQLLPLT-HLSLLPHPLMLELKKIQERKRKIITRSKKKE 460
Query: 1142 EQLSKKRRREERREK 1156
L + +R+ERR +
Sbjct: 461 ITLGTRIKRKERRSR 505
>TC12137 similar to UP|Q8L7W3 (Q8L7W3) At2g29210/F16P2.41, partial (9%)
Length = 579
Score = 33.1 bits (74), Expect = 0.26
Identities = 17/43 (39%), Positives = 30/43 (69%)
Frame = +3
Query: 1126 KENEKRKAHEAEKAKEEQLSKKRRREERREKYRTQDKSSKKMR 1168
KE +++K E + KEE K+RRREERR +R +++ ++K++
Sbjct: 51 KETQRKKREEKKLRKEE---KRRRREERR--HRREERRAEKLK 164
Score = 29.6 bits (65), Expect = 2.9
Identities = 17/46 (36%), Positives = 28/46 (59%), Gaps = 1/46 (2%)
Frame = +3
Query: 1119 KMKKRKLKENEKRKAHEAEKAKEEQLSKKRRREERR-EKYRTQDKS 1163
K +RK +E +K + E + +EE ++ RREERR EK + + K+
Sbjct: 51 KETQRKKREEKKLRKEEKRRRREE---RRHRREERRAEKLKLKSKT 179
>TC10427 similar to UP|Q9AU06 (Q9AU06) NADPH-cytochrome P450 oxydoreductase
isoform 3, partial (17%)
Length = 565
Score = 32.3 bits (72), Expect = 0.45
Identities = 26/71 (36%), Positives = 33/71 (45%), Gaps = 2/71 (2%)
Frame = -3
Query: 526 GASSDSESLNEEEEDDVMGNVSKWKESLAERTLSQKTPSL--MQLVYGESTVNLISINKE 583
G S S SL+ EE+ D+ G V SL + L L + LV G S N E
Sbjct: 464 GESFSSTSLSTEEDGDLSGTVVNL--SLVAKLLDDALGRLDGLSLVTGGSAPN------E 309
Query: 584 NDSSEDEESDG 594
ND + DE+ DG
Sbjct: 308 NDDASDEDGDG 276
>TC16669 similar to UP|Q9SLT9 (Q9SLT9) ZCF37 protein (T30E16.15), partial
(13%)
Length = 667
Score = 32.0 bits (71), Expect = 0.59
Identities = 28/109 (25%), Positives = 47/109 (42%), Gaps = 5/109 (4%)
Frame = +3
Query: 320 PCPLPSAAKKKGLRDKEKLFYAPMSGLGDLLYDKD----AVYININDHFVQFTKVDDENS 375
PC P +K+K RDK + +LL D D VY +N H + F + +S
Sbjct: 246 PCSSPVKSKRKENRDKNPFSSRGLDKFSELLEDLDQKRQKVYSRMNPHDISFVRFAYTSS 425
Query: 376 DITRKGNERDIGEVLVR-SLQETKYPINEKLENSMINLFAQSSEALAEA 423
+ D ++V+ ++ K NE+ + + F++S E A A
Sbjct: 426 N--------DFVPIVVKVKNRDHKKQGNEEFKVRHLTSFSESMEKTAAA 548
>TC17157 similar to UP|CAD27462 (CAD27462) Nucleosome assembly protein
1-like protein 3, partial (81%)
Length = 1298
Score = 32.0 bits (71), Expect = 0.59
Identities = 23/79 (29%), Positives = 35/79 (44%), Gaps = 8/79 (10%)
Frame = +2
Query: 471 EDDAAGDKSTNKDHLEEHIEFHNGRRRRRAI-------FGNDADQSDLMD-SSGNEDGSA 522
+DD D +L EH ++ G R I F +A+QSD D +EDG
Sbjct: 623 DDDIDDDAVEELQNLMEH-DYDIGSTIRDKIIPHAVSWFTGEAEQSDFEDIEEDDEDGDE 799
Query: 523 SDDGASSDSESLNEEEEDD 541
+D D E ++++DD
Sbjct: 800 DEDEDDDDDEEEEDDDDDD 856
>TC19663 similar to UP|Q8H6S8 (Q8H6S8) Translation initiation factor, partial
(10%)
Length = 485
Score = 31.6 bits (70), Expect = 0.77
Identities = 15/49 (30%), Positives = 30/49 (60%), Gaps = 4/49 (8%)
Frame = +3
Query: 1121 KKRKLKENEKRKAHEAEKAKEEQLSKK----RRREERREKYRTQDKSSK 1165
+++K +E EK + E E+ ++E+L ++ RRR++ REK + K +
Sbjct: 246 ERKKREEEEKLRKEEEERRRQEELERQAEEARRRKKEREKEKLLKKKQE 392
>TC11276 weakly similar to UP|HDA1_CHICK (P56517) Histone deacetylase 1 (HD1),
partial (5%)
Length = 636
Score = 31.6 bits (70), Expect = 0.77
Identities = 17/52 (32%), Positives = 30/52 (57%), Gaps = 1/52 (1%)
Frame = +1
Query: 1119 KMKKRKLKEN-EKRKAHEAEKAKEEQLSKKRRREERREKYRTQDKSSKKMRR 1169
K K+ + K+N +KRK + + + K+RRR+ R+K R + K K+ +R
Sbjct: 31 KRKRNQRKKNPKKRKRRKMSQRVKNPRKKRRRRKSPRKKKRRKMKRKKRRKR 186
>TC8053 similar to UP|Q94A20 (Q94A20) AT4g15470/dl3775w, partial (82%)
Length = 1109
Score = 31.6 bits (70), Expect = 0.77
Identities = 18/41 (43%), Positives = 26/41 (62%)
Frame = -3
Query: 581 NKENDSSEDEESDGDFFEPIEEVKKKNVRDGLNDGNVNTED 621
N++++ EDEE + P EEV K++V DG DGN + ED
Sbjct: 354 NQKDEGEEDEEKEERRIAPEEEV-KRSVEDG-EDGNESGED 238
>TC19988 similar to UP|CRT3_ARATH (O04153) Calreticulin 3 precursor, partial
(26%)
Length = 642
Score = 30.8 bits (68), Expect = 1.3
Identities = 17/52 (32%), Positives = 30/52 (57%), Gaps = 1/52 (1%)
Frame = +2
Query: 1119 KMKKRKLKENEK-RKAHEAEKAKEEQLSKKRRREERREKYRTQDKSSKKMRR 1169
+++K +E EK RKA E E+A+ + +RRR+ER +D+ + R+
Sbjct: 221 EIEKEAFEEEEKIRKAKEEEEAQRAREEGERRRKERGHDRHHRDRHRDRYRK 376
>BP062078
Length = 419
Score = 28.1 bits (61), Expect = 8.5
Identities = 20/74 (27%), Positives = 28/74 (37%), Gaps = 10/74 (13%)
Frame = -1
Query: 665 EEENDDVYGDF----------EDLETGEKIENNQTDDATHNGDDLEAETRRLKKLALQED 714
++END+ DF ED E E EN D++ G D+ E + QE
Sbjct: 380 DDENDEFEEDFETELNGGKHEEDEEVEEDAENENIDNSWSIGGDIRMEKYERSRGRCQEG 201
Query: 715 GGNENEAKFRRGQP 728
E G+P
Sbjct: 200 DHENXELLVNWGEP 159
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.314 0.133 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,800,753
Number of Sequences: 28460
Number of extensions: 216209
Number of successful extensions: 1158
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 1092
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1134
length of query: 1172
length of database: 4,897,600
effective HSP length: 100
effective length of query: 1072
effective length of database: 2,051,600
effective search space: 2199315200
effective search space used: 2199315200
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0122.3


