

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0119.8
(520 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
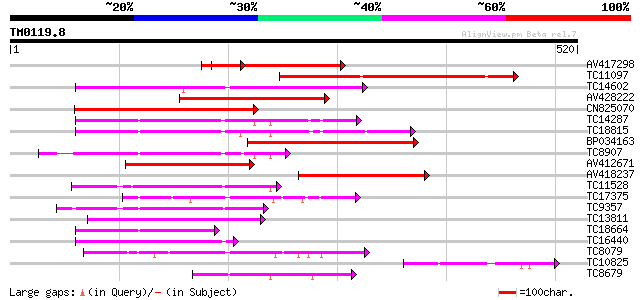
Score E
Sequences producing significant alignments: (bits) Value
AV417298 194 3e-50
TC11097 similar to UP|P93520 (P93520) Calcium/calmodulin-depende... 187 4e-48
TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinas... 186 1e-47
AV428222 183 5e-47
CN825070 162 9e-41
TC14287 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein k... 162 1e-40
TC18815 similar to UP|Q8W1D5 (Q8W1D5) CBL-interacting protein ki... 157 3e-39
BP034163 155 2e-38
TC8907 similar to UP|Q8H0X3 (Q8H0X3) Serine/threonine protein ki... 134 4e-32
AV412671 130 7e-31
AV418237 119 2e-27
TC11528 homologue to UP|Q39868 (Q39868) Protein kinase , partial... 109 1e-24
TC17375 mitogen-activated kinase kinase kinase alpha [Lotus corn... 96 2e-20
TC9357 homologue to UP|Q39193 (Q39193) Protein kinase (Protein k... 93 9e-20
TC13811 homologue to UP|O24431 (O24431) Calmodulin-like domain p... 92 2e-19
TC18664 similar to UP|Q944I6 (Q944I6) At2g30360/T9D9.17, partial... 90 8e-19
TC16440 similar to UP|Q93Y18 (Q93Y18) SNF1 related protein kinas... 89 2e-18
TC8079 homologue to UP|Q9M6S1 (Q9M6S1) MAP kinase 3, complete 83 1e-16
TC10825 similar to GB|AAP68339.1|31711966|BT008900 At1g74740 {Ar... 81 5e-16
TC8679 homologue to UP|Q43466 (Q43466) Protein kinase 3 , partia... 78 3e-15
>AV417298
Length = 424
Score = 194 bits (493), Expect = 3e-50
Identities = 98/123 (79%), Positives = 102/123 (82%)
Frame = +1
Query: 186 MKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVAPEVLKRKSGPQSDVW 245
+KP F FK R + LS GKKF DIVGSAYYVAPEVLKRKSGP+SDVW
Sbjct: 55 IKPRRFSFKGYRFWFV*FHKTWSLSLKTISGKKFQDIVGSAYYVAPEVLKRKSGPESDVW 234
Query: 246 SIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKDFVKKLLIKDPR 305
SIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKDFVKKLL+KDPR
Sbjct: 235 SIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKDFVKKLLVKDPR 414
Query: 306 ARL 308
ARL
Sbjct: 415 ARL 423
Score = 79.7 bits (195), Expect = 1e-15
Identities = 37/40 (92%), Positives = 37/40 (92%)
Frame = +2
Query: 177 HLRGLVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPG 216
HL GLVHRDMKPENFLFKS REDS LKATDFGLSDFIKPG
Sbjct: 2 HLHGLVHRDMKPENFLFKSNREDSALKATDFGLSDFIKPG 121
>TC11097 similar to UP|P93520 (P93520) Calcium/calmodulin-dependent protein
kinase homolog|CaM kinase homolog|MCK1, partial (36%)
Length = 949
Score = 187 bits (475), Expect = 4e-48
Identities = 98/219 (44%), Positives = 148/219 (66%)
Frame = +2
Query: 248 GVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKDFVKKLLIKDPRAR 307
GVITYILLCG RPFW +TE GIF+ VLR P+F PWP+++ AKDFVK+LL KD R R
Sbjct: 20 GVITYILLCGSRPFWARTESGIFRAVLRADPNFDDLPWPSVTPEAKDFVKRLLNKDYRKR 199
Query: 308 LTAAQALSHPWVREGGEASEIPIDISVLNNLRQFVTYSRFKQFALRALASILNEEELADL 367
+TAAQAL+HPW+R+ ++ IP+DI + ++ ++ + FK+ A++AL+ L EE+L L
Sbjct: 200 MTAAQALTHPWLRD--DSRPIPLDILIYKLVKSYLHATPFKRAAVKALSKALTEEQLVYL 373
Query: 368 KDQFDAIDVDKNGSISLEEMRQALAKDLSWKLKESRVLEILQAIDSNTDGLVDFTEFVAA 427
+ QF ++ +++G ISL+ + ALA+ + ++ESRVL+I+ ++ +DF EF AA
Sbjct: 374 RAQFRLLEPNRDGHISLDNFKMALARHATDAMRESRVLDIIHMMEPLAYRKMDFEEFCAA 553
Query: 428 TLHVHQLEEHDSDKWQLRSQAAFEKFDLDKDGYITPEEL 466
+QLE D+W+ + AAFE F+ + + I+ EEL
Sbjct: 554 ATSTYQLEA--LDRWEDIASAAFEHFEREGNRVISIEEL 664
>TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinase, complete
Length = 1256
Score = 186 bits (471), Expect = 1e-47
Identities = 101/272 (37%), Positives = 160/272 (58%), Gaps = 4/272 (1%)
Frame = +2
Query: 61 RYSLGKLLGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVKQEVNILKAVAGH 120
+Y L + +G G+FG + S D AVK ++KS + ++ E + ++ H
Sbjct: 65 QYQLCEEIGRGRFGTIFRCFHPISTDTFAVKLIDKSLLADSTDRHCLENEPKYMSLLSPH 244
Query: 121 ENVVQFYNAFDDDSYVYIVMELCEGGELLDRILANRYT---EKDAAVVVRQMLKVAAECH 177
N++Q ++ F+DD + +V+ELC+ LLDRI+A T E +AA +++Q+L+ A CH
Sbjct: 245 PNILQIFDVFEDDDVLSMVIELCQPLTLLDRIVAANGTSIPEVEAAGLMKQLLEAVAHCH 424
Query: 178 LRGLVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVAPEVLK-R 236
G+ HRD+KP+N LF + LK DFG +++ G++ +VG+ YYVAPEVL R
Sbjct: 425 RLGVAHRDVKPDNVLFGGGGD---LKLADFGSAEWFGDGRRMSGVVGTPYYVAPEVLMGR 595
Query: 237 KSGPQSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKDFV 296
+ G + DVWS GVI YI+L G PF+ + IF+ V+R F + + +S AAKD +
Sbjct: 596 EYGEKVDVWSCGVILYIMLSGTPPFYGDSAAEIFEAVIRGNLRFPSRIFRNVSPAAKDLL 775
Query: 297 KKLLIKDPRARLTAAQALSHPWVREGGEASEI 328
+K++ +DP R++A QAL HPW G+ +
Sbjct: 776 RKMICRDPSNRISAEQALRHPWFLSAGDTVNV 871
>AV428222
Length = 416
Score = 183 bits (465), Expect = 5e-47
Identities = 85/138 (61%), Positives = 104/138 (74%)
Frame = +3
Query: 156 RYTEKDAAVVVRQMLKVAAECHLRGLVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKP 215
RY E DA ++ Q+L V A CHL G+VHRD+KPENFLF S DS +K DFGLSDF++P
Sbjct: 3 RYPEDDAKAILLQILNVVAFCHLHGVVHRDLKPENFLFVSKEADSVMKVIDFGLSDFVRP 182
Query: 216 GKKFHDIVGSAYYVAPEVLKRKSGPQSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLR 275
++ +DIVGSAYYVAPEVL R ++D+WSIGVI+YILLCG RPFW +TE GIF+ VLR
Sbjct: 183 DQRLNDIVGSAYYVAPEVLHRSYSVEADLWSIGVISYILLCGSRPFWARTESGIFRSVLR 362
Query: 276 NKPDFRRKPWPTISNAAK 293
P+F PWP+IS AK
Sbjct: 363 ANPNFDDSPWPSISPEAK 416
>CN825070
Length = 634
Score = 162 bits (411), Expect = 9e-41
Identities = 78/170 (45%), Positives = 117/170 (67%), Gaps = 1/170 (0%)
Frame = +3
Query: 60 ERYSLGKLLGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVKQEVNILKAVAG 119
E Y+ G+ LG GQFG TY+ ++G A K + K K++ +DV +E+ I+ ++
Sbjct: 123 EVYTFGRKLGQGQFGTTYLCRHNSTGCTFACKSIPKRKLLCKEDYDDVWREIQIMHHLSE 302
Query: 120 HENVVQFYNAFDDDSYVYIVMELCEGGELLDRILAN-RYTEKDAAVVVRQMLKVAAECHL 178
H +VV+ + ++D + V+IVME+CEGGEL DRI+ +Y+E++AA ++R +++V CH
Sbjct: 303 HPHVVRIHGTYEDAASVHIVMEICEGGELFDRIVQKGQYSEREAAKLIRTIVEVVEACHS 482
Query: 179 RGLVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAYY 228
G++HRD+KPENFLF + ED+ LK TDFGLS F KPG+ F D+VGS YY
Sbjct: 483 LGVMHRDLKPENFLFDTVEEDAKLKTTDFGLSVFYKPGETFGDVVGSPYY 632
>TC14287 similar to UP|Q9LEU7 (Q9LEU7) Serine/threonine protein kinase-like
protein (CBL-interacting protein kinase 5), partial
(75%)
Length = 1999
Score = 162 bits (410), Expect = 1e-40
Identities = 92/267 (34%), Positives = 148/267 (54%), Gaps = 5/267 (1%)
Frame = +3
Query: 61 RYSLGKLLGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVKQEVNILKAVAGH 120
+Y +G++LG G F Y G + A+ + VA+K ++K K+ V+ +K+EV++++ V H
Sbjct: 429 KYEMGRVLGQGNFAKVYHGRNLATNENVAIKVIKKEKLKKDRLVKQIKREVSVMRLVR-H 605
Query: 121 ENVVQFYNAFDDDSYVYIVMELCEGGELLDRILANRYTEKDAAVVVRQMLKVAAECHLRG 180
++V+ +++VME +GGEL ++ + E DA +Q++ CH RG
Sbjct: 606 PHIVELKEVMATKGKIFLVMEYVKGGELFTKVNKGKLNEDDARKYFQQLISAVDFCHSRG 785
Query: 181 LVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIV---GSAYYVAPEVLKRK 237
+ HRD+KPEN L E+ LK +DFGLS + + +V G+ YVAPEVLK+K
Sbjct: 786 VTHRDLKPENLLLD---ENEDLKVSDFGLSALPEQRRDDGMLVTPCGTPAYVAPEVLKKK 956
Query: 238 --SGPQSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKDF 295
G ++D+WS GVI Y LL G PF + I+++ K ++ W IS AK+
Sbjct: 957 GYDGSKADIWSCGVILYALLSGYLPFQGENVMRIYRKAF--KAEYEFPEW--ISPQAKNL 1124
Query: 296 VKKLLIKDPRARLTAAQALSHPWVREG 322
+ LL+ DP R + + +S PW + G
Sbjct: 1125ISNLLVADPEKRYSIPEIISDPWFQYG 1205
>TC18815 similar to UP|Q8W1D5 (Q8W1D5) CBL-interacting protein kinase
CIPK25, partial (58%)
Length = 979
Score = 157 bits (398), Expect = 3e-39
Identities = 99/318 (31%), Positives = 165/318 (51%), Gaps = 6/318 (1%)
Frame = +2
Query: 61 RYSLGKLLGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVKQEVNILKAVAGH 120
+Y +G++LG G Y + SG+ VA+K + K+++ ++ +K+E++I++ V H
Sbjct: 35 KYEMGRVLGKGTLAKVYFAKEITSGEGVAIKVMSKARIKKEGMMDQIKREISIMRLVR-H 211
Query: 121 ENVVQFYNAFDDDSYVYIVMELCEGGELLDRILANRYTEKDAAVVVRQMLKVAAECHLRG 180
N+V + ++ +ME GGEL ++ + + A +Q++ CH RG
Sbjct: 212 PNIVNLKEVMATKTKIFFIMEYIRGGELFAKVAKGKLKDDLARRYFQQLISAVDYCHSRG 391
Query: 181 LVHRDMKPENFLFKSTREDSPLKATDFGLS---DFIKPGKKFHDIVGSAYYVAPEVLKRK 237
+ HRD+KPEN L E+ LK +DFGLS + ++ H G+ YVAPEVL++K
Sbjct: 392 VSHRDLKPENLLLD---ENENLKVSDFGLSGLPEQLRQDGLLHTQCGTPAYVAPEVLRKK 562
Query: 238 --SGPQSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKPDFRRKPWPTISNAAKDF 295
G ++D WS GVI Y LL G PF + ++ +VLR +F+ PW S +K
Sbjct: 563 GYDGFKTDTWSCGVILYALLAGCLPFQHENLMTMYNKVLR--AEFQFPPW--FSPESKKL 730
Query: 296 VKKLLIKDPRARLTAAQALSHPWVREGGEASEIPIDISVLNNLRQFVTYSRFKQFALRAL 355
+ K+L+ DP R+T + + W ++G AS IPI +N + S + + A
Sbjct: 731 ISKILVADPNRRITISSIMRVSWFQKGFSAS-IPIPDPDESNFNSDLNSSSEQSTKVVAA 907
Query: 356 ASILNE-EELADLKDQFD 372
A +N E ++ + FD
Sbjct: 908 AKFINAFEFISSMSSGFD 961
>BP034163
Length = 477
Score = 155 bits (392), Expect = 2e-38
Identities = 73/157 (46%), Positives = 104/157 (65%)
Frame = +1
Query: 219 FHDIVGSAYYVAPEVLKRKSGPQSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRNKP 278
F++I GS YY+APEVL+R GP+ D+WS GVI YILLCG PFW +TE + + ++R+
Sbjct: 4 FNEIXGSPYYMAPEVLRRHYGPEVDIWSAGVILYILLCGVPPFWAETEQVVAQAIIRSVV 183
Query: 279 DFRRKPWPTISNAAKDFVKKLLIKDPRARLTAAQALSHPWVREGGEASEIPIDISVLNNL 338
DF+R PWP +S+ AKD VKK+L DP+ RLTA + L HPW+ +A + + +V L
Sbjct: 184 DFKRDPWPKVSDNAKDLVKKMLNPDPKRRLTAQEVLDHPWLVHAKKAPNVSLGETVKARL 363
Query: 339 RQFVTYSRFKQFALRALASILNEEELADLKDQFDAID 375
+QF ++ K+ ALR +A L EE A L++ F +D
Sbjct: 364 KQFSVMNKLKKRALRVIAEHLTVEEAAGLREGFQGMD 474
>TC8907 similar to UP|Q8H0X3 (Q8H0X3) Serine/threonine protein kinase-like
protein, partial (44%)
Length = 941
Score = 134 bits (337), Expect = 4e-32
Identities = 79/238 (33%), Positives = 130/238 (54%), Gaps = 7/238 (2%)
Frame = +3
Query: 27 NRHQRQRAQRHVAGSRHVPSGMRADFGYDKDFDERYSLGKLLGHGQFGYTYVGVDKASGD 86
N QRQ+++ A R++ +Y +GK+LG G F Y G + + +
Sbjct: 276 NMDQRQQSRTMRAAKRNI-------------LFNKYEIGKILGQGNFAKVYHGRNMETNE 416
Query: 87 RVAVKRLEKSKMVLPAAVEDVKQEVNILKAVAGHENVVQFYNAFDDDSYVYIVMELCEGG 146
VA+K ++K ++ V+ +K+EV++++ V H ++V+ + +++V+E +GG
Sbjct: 417 SVAIKVIKKERLKKERLVKQIKREVSVMRLVR-HPHIVELKEVMATKTKIFMVVEYVKGG 593
Query: 147 ELLDRILANRYTEKDAAVVVRQMLKVAAECHLRGLVHRDMKPENFLFKSTREDSPLKATD 206
EL ++ + TE A +Q++ CH RG+ HRD+KPEN L ++ LK +D
Sbjct: 594 ELFAKLTKGKMTEVAARKYFQQLISAVDFCHSRGVTHRDLKPENLLLD---DNEDLKVSD 764
Query: 207 FGLSDFIKPGKKFHDIV-----GSAYYVAPEVLKRK--SGPQSDVWSIGVITYILLCG 257
FGLS P ++ D + G+ YVAPEVLK+K G ++D+WS GVI Y LLCG
Sbjct: 765 FGLSSL--PEQRRSDGMLLTPCGTPAYVAPEVLKKKGYDGSKADIWSCGVILYALLCG 932
>AV412671
Length = 359
Score = 130 bits (326), Expect = 7e-31
Identities = 61/119 (51%), Positives = 88/119 (73%), Gaps = 1/119 (0%)
Frame = +3
Query: 107 VKQEVNILKAVAGHENVVQFYNAFDDDSYVYIVMELCEGGELLDRILAN-RYTEKDAAVV 165
V++EV I++ + H N+V + ++DD+ V++VMELCEGGEL DRI+A YTE+ AA V
Sbjct: 3 VRREVEIMRHLPRHPNIVWLKDTYEDDNAVHLVMELCEGGELFDRIVARGHYTERAAAAV 182
Query: 166 VRQMLKVAAECHLRGLVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVG 224
+ +++V CH G++HRD+KPENFLF + +E + LKA DFGLS F KPG+ F++IVG
Sbjct: 183 TKTIVEVVQMCHKHGVMHRDLKPENFLFANKKETAALKAIDFGLSVFFKPGETFNEIVG 359
>AV418237
Length = 361
Score = 119 bits (297), Expect = 2e-27
Identities = 52/120 (43%), Positives = 85/120 (70%)
Frame = +2
Query: 266 EDGIFKEVLRNKPDFRRKPWPTISNAAKDFVKKLLIKDPRARLTAAQALSHPWVREGGEA 325
E GIF+++L + DF+ +PWP+IS++AKD ++K+L ++P+ RLTA Q L HPW+ + A
Sbjct: 2 EQGIFRQILMGRLDFQSEPWPSISDSAKDLIRKMLDRNPKTRLTAHQVLCHPWIVDDNIA 181
Query: 326 SEIPIDISVLNNLRQFVTYSRFKQFALRALASILNEEELADLKDQFDAIDVDKNGSISLE 385
+ P+D +VL+ L+QF ++ K+ ALR +A L+EEE+ LK+ F ID D +G+I+ +
Sbjct: 182 PDKPLDSAVLSRLKQFSAMNKLKKMALRVIAESLSEEEIGGLKELFKMIDADNSGTITFD 361
>TC11528 homologue to UP|Q39868 (Q39868) Protein kinase , partial (56%)
Length = 640
Score = 109 bits (272), Expect = 1e-24
Identities = 74/197 (37%), Positives = 106/197 (53%), Gaps = 4/197 (2%)
Frame = +3
Query: 57 DFDERYSLGKLLGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVKQEVNILKA 116
D +ERY K LG G FG + DK +G+ VAVK +E+ K + E+V++E+ I
Sbjct: 63 DMEERYEPLKELGSGNFGVARLARDKNTGELVAVKYIERGKKI----DENVQREI-INHR 227
Query: 117 VAGHENVVQFYNAFDDDSYVYIVMELCEGGELLDRIL-ANRYTEKDAAVVVRQMLKVAAE 175
H N+++F +++ IV+E GGEL +RI A R++E +A +Q++ +
Sbjct: 228 SLRHPNIIRFKEVLLTPTHLAIVLEYASGGELFERICSAGRFSEDEARYFFQQLISGVSY 407
Query: 176 CHLRGLVHRDMKPENFLFKSTREDSP-LKATDFGLSDFIKPGKKFHDIVGSAYYVAPEVL 234
CH + HRD+K EN L SP LK DFG S + VG+ Y+APEVL
Sbjct: 408 CHSMEICHRDLKLENTLLDG--NPSPRLKICDFGYSKSAILHSQPKSTVGTPAYIAPEVL 581
Query: 235 KRK--SGPQSDVWSIGV 249
RK G +DVWS GV
Sbjct: 582 SRKEYDGKVADVWSCGV 632
>TC17375 mitogen-activated kinase kinase kinase alpha [Lotus corniculatus
var. japonicus]
Length = 1422
Score = 95.5 bits (236), Expect = 2e-20
Identities = 70/224 (31%), Positives = 114/224 (50%), Gaps = 6/224 (2%)
Frame = +3
Query: 104 VEDVKQEVNILKAVAGHENVVQFYNAFDDDSYVYIVMELCEGGELLDRILANRYTEKDAA 163
++ + QE+N+L + H N+VQ+Y + + + + +E GG + ++L K+
Sbjct: 63 LKQLNQEINLLNQFS-HPNIVQYYGSELGEESLSVYLEYVSGGSI-HKLLQEYGAFKEPV 236
Query: 164 V--VVRQMLKVAAECHLRGLVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHD 221
+ RQ++ A H R VHRD+K N L E +K DFG+S I
Sbjct: 237 IQNYTRQIVSGLAYLHSRNTVHRDIKGANILVDPNGE---IKLADFGMSKHINSAASMLS 407
Query: 222 IVGSAYYVAPEVLKRKSGP--QSDVWSIGVITYILLCGRRPFWDKTED--GIFKEVLRNK 277
GS Y++APEV+ +G D+ S+G T + + +P W + E IFK + N
Sbjct: 408 FKGSPYWMAPEVVMNTNGYGLPVDISSLGC-TILEMATSKPPWSQFEGVAAIFK--IGNS 578
Query: 278 PDFRRKPWPTISNAAKDFVKKLLIKDPRARLTAAQALSHPWVRE 321
D P +S+ AK+F+K+ L +DP AR TA L+HP++R+
Sbjct: 579 KDMPEIP-EHLSDDAKNFIKQCLQRDPLARPTAQSLLNHPFIRD 707
>TC9357 homologue to UP|Q39193 (Q39193) Protein kinase (Protein kinase,
41K) , partial (71%)
Length = 1181
Score = 93.2 bits (230), Expect = 9e-20
Identities = 66/195 (33%), Positives = 101/195 (50%), Gaps = 1/195 (0%)
Frame = +1
Query: 44 VPSGMRADFGYDKDFDERYSLGKLLGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAA 103
V GM +D D RY L + +G G FG + DK + + VAVK +E+ +
Sbjct: 316 VGPGMDMPIMHDSD---RYDLVRDIGSGNFGVARLMQDKQTKELVAVKYIERGDKI---- 474
Query: 104 VEDVKQEVNILKAVAGHENVVQFYNAFDDDSYVYIVMELCEGGELLDRIL-ANRYTEKDA 162
E+VK+E+ +++ H N+V+F +++ IVME GGEL +RI A R+TE +A
Sbjct: 475 DENVKREIINHRSLR-HPNIVRFKEVILTPTHLAIVMEYASGGELFERICNAGRFTEDEA 651
Query: 163 AVVVRQMLKVAAECHLRGLVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDI 222
+Q++ + CH + HRD+K EN L + LK DFG S +
Sbjct: 652 RFFFQQLISGVSYCHAMQVCHRDLKLENTLLDGS-PTPRLKICDFGYSKSSVLHSQPKST 828
Query: 223 VGSAYYVAPEVLKRK 237
VG+ Y+APEVL ++
Sbjct: 829 VGTPAYIAPEVLSKQ 873
>TC13811 homologue to UP|O24431 (O24431) Calmodulin-like domain protein
kinase isoenzyme gamma , partial (27%)
Length = 573
Score = 92.4 bits (228), Expect = 2e-19
Identities = 55/164 (33%), Positives = 89/164 (53%), Gaps = 1/164 (0%)
Frame = +1
Query: 72 QFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVKQEVNILKAVAGHENVVQFYNAFD 131
QFG TY+ + ++ A K + K K+V + ED+K+E+ I++ + G N+V+F A++
Sbjct: 1 QFGVTYLCTEISTKLLYACKSISKRKLVSKSDKEDIKREIQIMQHLIGQPNIVEFKGAYE 180
Query: 132 DDSYVYIVMELCEGGELLDRILA-NRYTEKDAAVVVRQMLKVAAECHLRGLVHRDMKPEN 190
D V++VMELC GGEL DRI+A Y+E+ AA + RQ++ V CH G +
Sbjct: 181 DKQSVHVVMELCAGGELFDRIIAKGHYSERAAASICRQIVNVVHVCHFMGCDA*GSEARE 360
Query: 191 FLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVAPEVL 234
FL S + F L F G F+ ++ + + ++P L
Sbjct: 361 FLII*QG*KSTSQGHGFWLVCFH**G*IFYPLIDNLFPISPAFL 492
>TC18664 similar to UP|Q944I6 (Q944I6) At2g30360/T9D9.17, partial (34%)
Length = 643
Score = 90.1 bits (222), Expect = 8e-19
Identities = 49/133 (36%), Positives = 74/133 (54%), Gaps = 1/133 (0%)
Frame = +3
Query: 61 RYSLGKLLGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVKQEVNILKAVAGH 120
+Y +G+LLG G F Y + +G VAVK + K K++ VK+EV+I+ + H
Sbjct: 90 KYEVGRLLGCGAFAKVYHARNIETGQSVAVKVINKKKVIGTGLTGHVKREVSIMSRLR-H 266
Query: 121 ENVVQFYNAFDDDSYVYIVMELCEGGELLDRILAN-RYTEKDAAVVVRQMLKVAAECHLR 179
N+V+ + + +Y VME +GGEL RI R++E A +Q++ CH R
Sbjct: 267 PNIVRLHEVLATKTKIYFVMEFAKGGELFARISTKGRFSEDLARRYFQQLISAVGYCHSR 446
Query: 180 GLVHRDMKPENFL 192
G+ HRD+KPEN L
Sbjct: 447 GVFHRDLKPENLL 485
>TC16440 similar to UP|Q93Y18 (Q93Y18) SNF1 related protein kinase, partial
(30%)
Length = 549
Score = 88.6 bits (218), Expect = 2e-18
Identities = 47/151 (31%), Positives = 79/151 (52%), Gaps = 1/151 (0%)
Frame = +1
Query: 61 RYSLGKLLGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVKQEVNILKAVAGH 120
+Y L + LG G F Y V G VAVK ++KSK V + + +E++ ++ + H
Sbjct: 88 KYQLTRFLGRGNFAKVYQAVSLTDGTTVAVKMIDKSKTVDASMEPRIVREIDAMRRLQHH 267
Query: 121 ENVVQFYNAFDDDSYVYIVMELCEGGELLDRI-LANRYTEKDAAVVVRQMLKVAAECHLR 179
N+++ + + +Y++++ GGEL +I R E A +Q++ CH
Sbjct: 268 PNILKIHEVMATRTKIYLIVDYAGGGELFSKISRRGRLPEPLARRYFQQLVSALCFCHRN 447
Query: 180 GLVHRDMKPENFLFKSTREDSPLKATDFGLS 210
G+ HRD+KP+N L + + LK +DFGLS
Sbjct: 448 GVAHRDLKPQNLLLDA---EGNLKVSDFGLS 531
>TC8079 homologue to UP|Q9M6S1 (Q9M6S1) MAP kinase 3, complete
Length = 1550
Score = 82.8 bits (203), Expect = 1e-16
Identities = 78/299 (26%), Positives = 139/299 (46%), Gaps = 36/299 (12%)
Frame = +3
Query: 68 LGHGQFGYTYVGVDKASGDRVAVKRLEKSKMVLPAAVEDVKQEVNILKAVAGHENVVQFY 127
+G G +G ++ + + VAVK++ + A ++ E+ +L+ + HENV+
Sbjct: 309 IGRGAYGIVCSLLNTETNELVAVKKIANAFDNHMDAKRTLR-EIKLLRHL-DHENVIALR 482
Query: 128 NAFD-----DDSYVYIVMELCEGGELLDRILANR-YTEKDAAVVVRQMLKVAAECHLRGL 181
+ + + VYI EL + +L I +N+ +E+ + Q+L+ H +
Sbjct: 483 DVIPPPLRREFTDVYITTELMDT-DLHQIIRSNQGLSEEHCQYFLYQVLRGLKYIHSANI 659
Query: 182 VHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAYYVAPEVLKRKSGPQ 241
+HRD+KP N L S + LK DFGL+ + V + +Y APE+L S
Sbjct: 660 IHRDLKPSNLLLNSNCD---LKIIDFGLARPTVENDFMTEYVVTRWYRAPELLLNSSDYT 830
Query: 242 S--DVWSIGVITYILLCGRRPFWD-------------------KTEDGIFKE-----VLR 275
S DVWS+G I ++ L ++P + + + G+ K +R
Sbjct: 831 SAIDVWSVGCI-FMELMNKKPLFPGKDHVHQLRLLTELLGTPTEADLGLVKNDDVRRYIR 1007
Query: 276 NKPDFRRKP----WPTISNAAKDFVKKLLIKDPRARLTAAQALSHPWVREGGEASEIPI 330
P + R+P +P + A D V K+L DP R+T QAL+HP++ + + ++ P+
Sbjct: 1008QLPQYPRQPLTKVFPHVHPMAMDLVDKMLTIDPTKRITVEQALAHPYLEKLHDVADEPV 1184
>TC10825 similar to GB|AAP68339.1|31711966|BT008900 At1g74740 {Arabidopsis
thaliana;}, partial (32%)
Length = 1091
Score = 80.9 bits (198), Expect = 5e-16
Identities = 46/149 (30%), Positives = 84/149 (55%), Gaps = 6/149 (4%)
Frame = +1
Query: 362 EELADLKDQFDAIDVDKNGSISLEEMRQALAKDLSWKLKESRVLEILQAIDSNTDGLVDF 421
EE+ +KD F +D DK+G +S EE++ L K + +L + + +++ D + +G++D+
Sbjct: 4 EEVEIIKDMFTLMDTDKDGRVSYEELKAGLRK-VGSQLADQEIKMLMEVADVDGNGVLDY 180
Query: 422 TEFVAATLHVHQLEEHDSDKWQLRSQAAFEKFDLDKDGYITPEELRM----HTGLRGS-- 475
EFVA T+H+ ++E + AF+ FD D GYI EL+ +G+ +
Sbjct: 181 GEFVAVTIHLQKMENDE------HFHKAFKFFDKDGSGYIESGELQEALADESGVTDADV 342
Query: 476 IDPLLEEADIDKDGKISLSEFRRLLRTAS 504
++ ++ E D DKDG+IS EF +++ +
Sbjct: 343 LNDIMREVDTDKDGRISYEEFVAMMKAGT 429
>TC8679 homologue to UP|Q43466 (Q43466) Protein kinase 3 , partial (70%)
Length = 972
Score = 78.2 bits (191), Expect = 3e-15
Identities = 46/155 (29%), Positives = 76/155 (48%), Gaps = 4/155 (2%)
Frame = +2
Query: 168 QMLKVAAECHLRGLVHRDMKPENFLFKSTREDSPLKATDFGLSDFIKPGKKFHDIVGSAY 227
Q++ CH + HRD+K EN L + LK DFG S + VG+
Sbjct: 5 QLISGVHYCHALQICHRDLKLENTLLDGSPAPR-LKICDFGYSKSSLLHSRPKSTVGTPA 181
Query: 228 YVAPEVLKRK--SGPQSDVWSIGVITYILLCGRRPFWDKTEDGIFKEVLRN--KPDFRRK 283
Y+APEVL R+ G +DVWS GV Y++L G PF D + F++ ++ ++
Sbjct: 182 YIAPEVLSRREYDGKLADVWSCGVTLYVMLVGAYPFEDHDDPRNFRKTIQRIMAVQYKIP 361
Query: 284 PWPTISNAAKDFVKKLLIKDPRARLTAAQALSHPW 318
+ IS + + ++ + +P R++ + +HPW
Sbjct: 362 DYVHISQDCRHLLSRIFVANPLRRISLKEIKNHPW 466
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.135 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,693,661
Number of Sequences: 28460
Number of extensions: 91879
Number of successful extensions: 821
Number of sequences better than 10.0: 192
Number of HSP's better than 10.0 without gapping: 738
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 747
length of query: 520
length of database: 4,897,600
effective HSP length: 94
effective length of query: 426
effective length of database: 2,222,360
effective search space: 946725360
effective search space used: 946725360
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0119.8


