

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0118c.15
(602 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
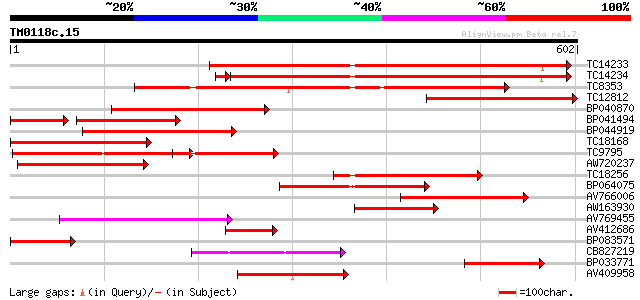
Score E
Sequences producing significant alignments: (bits) Value
TC14233 homologue to UP|Q41027 (Q41027) PsHSC71.0, partial (67%) 473 e-134
TC14234 homologue to UP|Q41027 (Q41027) PsHSC71.0, partial (66%) 447 e-127
TC8353 homologue to UP|HS7M_PHAVU (Q01899) Heat shock 70 kDa pro... 369 e-102
TC12812 weakly similar to UP|Q8RVV9 (Q8RVV9) Heat shock protein ... 314 2e-86
BP040870 241 2e-64
BP041494 165 2e-63
BP044919 228 2e-60
TC18168 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous heat... 225 2e-59
TC9795 homologue to UP|Q9ATB8 (Q9ATB8) BiP-isoform D (Fragment),... 220 6e-58
AW720237 193 8e-50
TC18256 homologue to UP|Q9M4E8 (Q9M4E8) Heat shock protein 70, p... 181 3e-46
BP064075 178 3e-45
AV766006 139 1e-33
AW163930 137 5e-33
AV769455 117 7e-27
AV412686 113 1e-25
BP083571 101 4e-22
CB827219 90 9e-19
BP033771 88 3e-18
AV409958 82 3e-16
>TC14233 homologue to UP|Q41027 (Q41027) PsHSC71.0, partial (67%)
Length = 1570
Score = 473 bits (1218), Expect = e-134
Identities = 246/398 (61%), Positives = 308/398 (76%), Gaps = 14/398 (3%)
Frame = +1
Query: 213 VSLVTIDEGIFKVNATVGDTYLGGVDFDNNLVNFLVDFFQRKHNKDITENVKSLRRLRLA 272
VSL+TI+EGIF+V AT GDT+LGG DFDN +VN V F+RK+ KDI+ N ++LRRLR A
Sbjct: 1 VSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTA 180
Query: 273 CEKAKRILSSTSQTTIELDSLCGGIDLHVTVTQGLFEKLNKDLFEKCMEIVEKCLVEAKI 332
CE+AKR LSST+QTTIE+DSL GID + +T+ FE++N DLF KCME VEKCL +AK+
Sbjct: 181 CERAKRTLSSTAQTTIEIDSLYEGIDFYSPITRARFEEMNMDLFRKCMEPVEKCLRDAKM 360
Query: 333 GKSEVHELVLVGGSTRIPKVQQLLKEMFSVHGRVKELCKSINPDEAVAYGAAVQAAILSS 392
K VH++VLVGGSTRIPKVQQLL++ F+ KELCKSINPDEAVAYGAAVQAAILS
Sbjct: 361 DKRSVHDVVLVGGSTRIPKVQQLLQDFFNG----KELCKSINPDEAVAYGAAVQAAILSG 528
Query: 393 EGDKKVEELLLLDVMPLSLGIEVVEGVMSVLIPKNTTIPTKKERTFRTTSDNQTSVWIKV 452
EG++KV++LLLLDV PLSLG+E GVM+VLIP+NTTIPTKKE+ F T SDNQ V I+V
Sbjct: 529 EGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQV 708
Query: 453 YEGEGAKTDKNFFLGKFELSGFSPAPKGDTEINVCFDVDADGIVEVSAEDMSLRLKKTIT 512
+EGE +T N LGKFELSG PAP+G +I VCFD+DA+GI+ VSAED + K IT
Sbjct: 709 FEGERTRTRDNNLLGKFELSGIPPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKIT 888
Query: 513 ITNRLGRLSQEEMRRMVRDAAQYKAEDEEVRKKVKAKNSLENYAYEARDRVK-------- 564
ITN GRLS++++ +MV++A +YK+EDEE +KKV+AKN+LENYAY R+ VK
Sbjct: 889 ITNDKGRLSKDDIEKMVQEAEKYKSEDEEHKKKVEAKNALENYAYNMRNTVKDEKIAGKL 1068
Query: 565 ------KLEKMVEEVIEWLDRNQLAEAEEFEYKKQELE 596
K+E +E+ I+WL+ NQLAEA+EFE K +ELE
Sbjct: 1069DSADKTKIEDAIEQAIQWLESNQLAEADEFEDKMKELE 1182
>TC14234 homologue to UP|Q41027 (Q41027) PsHSC71.0, partial (66%)
Length = 1528
Score = 447 bits (1149), Expect(2) = e-127
Identities = 233/376 (61%), Positives = 288/376 (75%), Gaps = 14/376 (3%)
Frame = +3
Query: 235 GGVDFDNNLVNFLVDFFQRKHNKDITENVKSLRRLRLACEKAKRILSSTSQTTIELDSLC 294
GG DFDN +VN V F+RK+ KDI+ N ++LRRLR ACE+AKR LSST+QTTIE+DSL
Sbjct: 48 GGEDFDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLY 227
Query: 295 GGIDLHVTVTQGLFEKLNKDLFEKCMEIVEKCLVEAKIGKSEVHELVLVGGSTRIPKVQQ 354
GID + +T+ FE+LN DLF KCME VEKCL +AK+ K VH++VLVGGSTRIPKVQQ
Sbjct: 228 EGIDFYSPITRARFEELNMDLFRKCMEPVEKCLRDAKMDKKSVHDVVLVGGSTRIPKVQQ 407
Query: 355 LLKEMFSVHGRVKELCKSINPDEAVAYGAAVQAAILSSEGDKKVEELLLLDVMPLSLGIE 414
LL++ F+ KELCKSINPDEAVAYGAAVQAAILS EG++KV++LLLLDV PLSLG+E
Sbjct: 408 LLQDFFNG----KELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLE 575
Query: 415 VVEGVMSVLIPKNTTIPTKKERTFRTTSDNQTSVWIKVYEGEGAKTDKNFFLGKFELSGF 474
GVM+VLIP+NTTIPTKKE+ F T SDNQ V I+VYEGE +T N LGKFELSG
Sbjct: 576 TAGGVMTVLIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERTRTRDNNLLGKFELSGI 755
Query: 475 SPAPKGDTEINVCFDVDADGIVEVSAEDMSLRLKKTITITNRLGRLSQEEMRRMVRDAAQ 534
PAP+G +I VCFD+DA+GI+ VSAED + K ITITN GRLS+E++ +MV++A +
Sbjct: 756 PPAPRGVPQITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEDIEKMVQEAEK 935
Query: 535 YKAEDEEVRKKVKAKNSLENYAYEARDRV--------------KKLEKMVEEVIEWLDRN 580
YK+EDEE +KKV+AKN+LENYAY R+ + KK+E +E+ I+WLD N
Sbjct: 936 YKSEDEEHKKKVEAKNALENYAYNMRNTIKDEKIGGKLDPADKKKIEDAIEQAIQWLDSN 1115
Query: 581 QLAEAEEFEYKKQELE 596
QLAEA+EFE K +ELE
Sbjct: 1116QLAEADEFEDKMKELE 1163
Score = 27.7 bits (60), Expect(2) = e-127
Identities = 11/16 (68%), Positives = 14/16 (86%)
Frame = +1
Query: 219 DEGIFKVNATVGDTYL 234
+EGIF+V AT GDT+L
Sbjct: 1 EEGIFEVKATAGDTHL 48
>TC8353 homologue to UP|HS7M_PHAVU (Q01899) Heat shock 70 kDa protein,
mitochondrial precursor, partial (74%)
Length = 1752
Score = 369 bits (946), Expect = e-102
Identities = 209/402 (51%), Positives = 269/402 (65%), Gaps = 4/402 (0%)
Frame = +3
Query: 133 EVAEAYLGHEVKRAVITVPAYFNNAQRQSTKDAGEIAGFDVMRIINEPTAASIAYGLDKK 192
E AEAYLG V +AVITVPAYFN+A RQ+TKDAG IAG DV RIINEPTAA+++YG+ K
Sbjct: 3 ETAEAYLGKSVSKAVITVPAYFNDAHRQATKDAGRIAGLDVQRIINEPTAAALSYGMTNK 182
Query: 193 GWRKGEQNVLVFDLGGGTFDVSLVTIDEGIFKVNATVGDTYLGGVDFDNNLVNFLVDFFQ 252
E + VFDLGGGTFDVS++ I G+F+V AT GDT+LGG DFDN L++FLVD F+
Sbjct: 183 -----EGLIAVFDLGGGTFDVSILEISNGVFEVKATNGDTFLGGEDFDNALLDFLVDEFK 347
Query: 253 RKHNKDITENVKSLRRLRLACEKAKRILSSTSQTTIELDSLC----GGIDLHVTVTQGLF 308
R + D++++ +L+RLR A EKAK LSSTSQT I L + G L++T+T+ F
Sbjct: 348 RTESIDLSKDKLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHLNITLTRSKF 527
Query: 309 EKLNKDLFEKCMEIVEKCLVEAKIGKSEVHELVLVGGSTRIPKVQQLLKEMFSVHGRVKE 368
E L L E+ + CL +A I EV E++LVGG TR+PKVQ+++ +F K
Sbjct: 528 EALVNHLIERTKAPCKNCLKDANISIKEVDEVLLVGGMTRVPKVQEVVSAIFG-----KS 692
Query: 369 LCKSINPDEAVAYGAAVQAAILSSEGDKKVEELLLLDVMPLSLGIEVVEGVMSVLIPKNT 428
K +NPDEAVA GAA+Q IL + V+ELLLLDV PLSLGIE + G+ + LI +NT
Sbjct: 693 PSKGVNPDEAVAMGAAIQGGILRGD----VKELLLLDVTPLSLGIETLGGIFTRLINRNT 860
Query: 429 TIPTKKERTFRTTSDNQTSVWIKVYEGEGAKTDKNFFLGKFELSGFSPAPKGDTEINVCF 488
TIPTKK + F T +DNQT V IKV +GE N LG+FEL G PAP+G +I V F
Sbjct: 861 TIPTKKSQVFSTAADNQTQVGIKVLQGEREMASDNKSLGEFELVGIPPAPRGLPQIEVTF 1040
Query: 489 DVDADGIVEVSAEDMSLRLKKTITITNRLGRLSQEEMRRMVR 530
D+DA+GIV VSA+D S ++ ITI L + RR ++
Sbjct: 1041DIDANGIVTVSAKDKSTGKEQQITIXGHLVVSQTMKSRRWLK 1166
>TC12812 weakly similar to UP|Q8RVV9 (Q8RVV9) Heat shock protein 70
(Fragment), partial (48%)
Length = 746
Score = 314 bits (805), Expect = 2e-86
Identities = 158/160 (98%), Positives = 158/160 (98%)
Frame = +1
Query: 443 DNQTSVWIKVYEGEGAKTDKNFFLGKFELSGFSPAPKGDTEINVCFDVDADGIVEVSAED 502
DNQTSVWIKVYEGEGAK DKNFFLGKFELSGFSPAPKGDTEINVCFDVDADGIVEVSAED
Sbjct: 1 DNQTSVWIKVYEGEGAKADKNFFLGKFELSGFSPAPKGDTEINVCFDVDADGIVEVSAED 180
Query: 503 MSLRLKKTITITNRLGRLSQEEMRRMVRDAAQYKAEDEEVRKKVKAKNSLENYAYEARDR 562
MSLRLKKTITITNRLGRLSQEEM RMVRDAAQYKAEDEEVRKKVKAKNSLENYAYEARDR
Sbjct: 181 MSLRLKKTITITNRLGRLSQEEMSRMVRDAAQYKAEDEEVRKKVKAKNSLENYAYEARDR 360
Query: 563 VKKLEKMVEEVIEWLDRNQLAEAEEFEYKKQELEKDMPFV 602
VKKLEKMVEEVIEWLDRNQLAEAEEFEYKKQELEKDMPFV
Sbjct: 361 VKKLEKMVEEVIEWLDRNQLAEAEEFEYKKQELEKDMPFV 480
>BP040870
Length = 505
Score = 241 bits (616), Expect = 2e-64
Identities = 117/167 (70%), Positives = 143/167 (85%)
Frame = +2
Query: 109 VTYKGEERKLAAEEISSMVLFKMKEVAEAYLGHEVKRAVITVPAYFNNAQRQSTKDAGEI 168
V YKGEE++ +AEEISSMVL KMKE+AEAYLG +K AV+TVPAYFN++QRQ+TKDAG I
Sbjct: 5 VNYKGEEKQFSAEEISSMVLMKMKEIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVI 184
Query: 169 AGFDVMRIINEPTAASIAYGLDKKGWRKGEQNVLVFDLGGGTFDVSLVTIDEGIFKVNAT 228
+G +VMRIINEPTAA+IAYGLDKK GE+NVL+FDLGGGTFDVSL+TI+EGIF+V +T
Sbjct: 185 SGLNVMRIINEPTAAAIAYGLDKKATSTGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKST 364
Query: 229 VGDTYLGGVDFDNNLVNFLVDFFQRKHNKDITENVKSLRRLRLACEK 275
GDT+LGG DFDN +VN V F+RK+ KDI+ N ++LRRLR ACE+
Sbjct: 365 AGDTHLGGEDFDNRMVNHFVQEFKRKNKKDISGNARALRRLRTACER 505
>BP041494
Length = 572
Score = 165 bits (418), Expect(2) = 2e-63
Identities = 80/110 (72%), Positives = 93/110 (83%)
Frame = +3
Query: 72 DAKRLIGRRFSDRSVQQDMKLWPFKVVQGTRDKPMISVTYKGEERKLAAEEISSMVLFKM 131
DAKRLIGR+FSD SV+ DMKLWPFKV+ G D+PMI V YK EE++ AEEISSMVL KM
Sbjct: 243 DAKRLIGRKFSDDSVESDMKLWPFKVISGPVDRPMIVVNYKNEEKQFTAEEISSMVLIKM 422
Query: 132 KEVAEAYLGHEVKRAVITVPAYFNNAQRQSTKDAGEIAGFDVMRIINEPT 181
+E+AEAYLG VK V+TVPAYFN++QRQ+TKDAG IAG +VMRIINEPT
Sbjct: 423 REIAEAYLGSTVKNVVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPT 572
Score = 94.7 bits (234), Expect(2) = 2e-63
Identities = 45/62 (72%), Positives = 51/62 (81%)
Frame = +1
Query: 1 MATSKKDKAIGIDLGTTYSCVAVWNHNRVEIIPNELGNRTTPSYVAFTDTERLMGDAAIN 60
M + AIGIDLGTTYSCV VW ++RVEIIPN+ GNRTTPSYVAFTD ERL+GDAA N
Sbjct: 61 MVGEGESPAIGIDLGTTYSCVGVWQNDRVEIIPNDQGNRTTPSYVAFTDFERLIGDAAKN 240
Query: 61 QL 62
Q+
Sbjct: 241 QM 246
>BP044919
Length = 519
Score = 228 bits (582), Expect = 2e-60
Identities = 112/164 (68%), Positives = 136/164 (82%)
Frame = +3
Query: 78 GRRFSDRSVQQDMKLWPFKVVQGTRDKPMISVTYKGEERKLAAEEISSMVLFKMKEVAEA 137
GR++SD +Q+D+KLWPFKV+ GT DKPMI V +KGEE+KL EEISSMVL KMKE+AEA
Sbjct: 27 GRKYSDSVIQEDVKLWPFKVIAGTDDKPMIVVKHKGEEKKLVPEEISSMVLSKMKEIAEA 206
Query: 138 YLGHEVKRAVITVPAYFNNAQRQSTKDAGEIAGFDVMRIINEPTAASIAYGLDKKGWRKG 197
+L VK AV+TVPAYFN+AQR++TKDAG IAG +VMRIINEPTAA++AYGL K+ G
Sbjct: 207 FLESSVKNAVVTVPAYFNDAQRKATKDAGTIAGLNVMRIINEPTAAALAYGLQKRANLVG 386
Query: 198 EQNVLVFDLGGGTFDVSLVTIDEGIFKVNATVGDTYLGGVDFDN 241
E+NV +FDLGGGTFDVSL+TI + F+V AT GDT+LG DFDN
Sbjct: 387 ERNVFIFDLGGGTFDVSLLTIKDDNFEVKATSGDTHLGXKDFDN 518
>TC18168 homologue to UP|Q8GSN4 (Q8GSN4) Non-cell-autonomous heat shock
cognate protein 70, partial (23%)
Length = 508
Score = 225 bits (573), Expect = 2e-59
Identities = 111/150 (74%), Positives = 126/150 (84%)
Frame = +2
Query: 1 MATSKKDKAIGIDLGTTYSCVAVWNHNRVEIIPNELGNRTTPSYVAFTDTERLMGDAAIN 60
MA + AIGIDLGTTYSCV VW H+RVEII N+ GNRTTPSYVAFTDTERL+GDAA N
Sbjct: 59 MAGKGEGPAIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDTERLIGDAAKN 238
Query: 61 QLALNPHNTVFDAKRLIGRRFSDRSVQQDMKLWPFKVVQGTRDKPMISVTYKGEERKLAA 120
Q+A+NP NTVFDAKRLIGRR SD SVQ DMKLWPFKV G +KPMI V+YKGE+++ AA
Sbjct: 239 QVAMNPINTVFDAKRLIGRRVSDASVQSDMKLWPFKVNSGPAEKPMIQVSYKGEDKQFAA 418
Query: 121 EEISSMVLFKMKEVAEAYLGHEVKRAVITV 150
EEISSMVL KM+E+AEAYLG +K AV+TV
Sbjct: 419 EEISSMVLMKMREIAEAYLGSTIKNAVVTV 508
>TC9795 homologue to UP|Q9ATB8 (Q9ATB8) BiP-isoform D (Fragment), partial
(63%)
Length = 981
Score = 220 bits (560), Expect = 6e-58
Identities = 113/194 (58%), Positives = 142/194 (72%), Gaps = 1/194 (0%)
Frame = +1
Query: 4 SKKDKAIGIDLGTTYSCVAVWNHNRVEIIPNELGNRTTPSYVAFTDTERLMGDAAINQLA 63
SK IGIDLGTTYSCV V+ + VEII N+ GNR TPS+VAF D ERL+G+AA NQ A
Sbjct: 136 SKLGTVIGIDLGTTYSCVGVYRNGHVEIIANDQGNRITPSWVAFNDGERLIGEAAKNQFA 315
Query: 64 LNPHNTVFDAKRLIGRRFSDRSVQQDMKLWPFKVVQGTRDKPMISVTYK-GEERKLAAEE 122
+NP T+FD KRLIGR+F D+ VQ+DMKL P+K+V KP I V K GE + + EE
Sbjct: 316 VNPERTIFDVKRLIGRKFEDKEVQRDMKLVPYKIV-NKDGKPYIQVKIKDGETKVFSPEE 492
Query: 123 ISSMVLFKMKEVAEAYLGHEVKRAVITVPAYFNNAQRQSTKDAGEIAGFDVMRIINEPTA 182
+S+M+L KMKE AEA+LG ++ AV+TVPAYFN+AQRQ+TKDAG IAG +V RIINEPT
Sbjct: 493 VSAMILTKMKETAEAFLGKKINDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTC 672
Query: 183 ASIAYGLDKKGWRK 196
+ ++ WR+
Sbjct: 673 CCHCLWIGQERWRE 714
Score = 126 bits (316), Expect = 1e-29
Identities = 61/113 (53%), Positives = 85/113 (74%)
Frame = +2
Query: 173 VMRIINEPTAASIAYGLDKKGWRKGEQNVLVFDLGGGTFDVSLVTIDEGIFKVNATVGDT 232
++ + P AA+IAYGLDKKG GE+N+LVFDLGGGTFDVS++TID G+F+V AT GDT
Sbjct: 644 LLELSMNPPAAAIAYGLDKKG---GEKNILVFDLGGGTFDVSILTIDNGVFEVLATNGDT 814
Query: 233 YLGGVDFDNNLVNFLVDFFQRKHNKDITENVKSLRRLRLACEKAKRILSSTSQ 285
+LGG DFD ++ + + ++KH KDI+++ ++L +LR E+AKR LSS Q
Sbjct: 815 HLGGEDFDQRIMEYFIKLIKKKHGKDISKDNRALGKLRREAERAKRALSSQHQ 973
>AW720237
Length = 459
Score = 193 bits (490), Expect = 8e-50
Identities = 96/139 (69%), Positives = 112/139 (80%)
Frame = +3
Query: 9 AIGIDLGTTYSCVAVWNHNRVEIIPNELGNRTTPSYVAFTDTERLMGDAAINQLALNPHN 68
AIGIDLGTTYSCV VW ++RVEII N+ GNRTTPSYV FT++ERL+GDAA NQ+A NP N
Sbjct: 42 AIGIDLGTTYSCVGVWMNDRVEIIANDQGNRTTPSYVGFTESERLIGDAAKNQVARNPIN 221
Query: 69 TVFDAKRLIGRRFSDRSVQQDMKLWPFKVVQGTRDKPMISVTYKGEERKLAAEEISSMVL 128
TVFDAKRLIGR+F D VQ+D+KLWPFKV G KP+I+V YKGE +K AE++SSMVL
Sbjct: 222 TVFDAKRLIGRKFDDPIVQKDIKLWPFKVEAGPDGKPLIAVKYKGETKKFHAEQVSSMVL 401
Query: 129 FKMKEVAEAYLGHEVKRAV 147
KMKE AEAYL +K V
Sbjct: 402 TKMKETAEAYLNKTIKDIV 458
>TC18256 homologue to UP|Q9M4E8 (Q9M4E8) Heat shock protein 70, partial
(24%)
Length = 476
Score = 181 bits (459), Expect = 3e-46
Identities = 96/159 (60%), Positives = 118/159 (73%)
Frame = +3
Query: 344 GGSTRIPKVQQLLKEMFSVHGRVKELCKSINPDEAVAYGAAVQAAILSSEGDKKVEELLL 403
GGSTRIPKVQQLLK+ F KE K +NPDEAVAYGAAVQ +ILS EG ++ +++LL
Sbjct: 3 GGSTRIPKVQQLLKDYFEG----KEPNKGVNPDEAVAYGAAVQGSILSGEGGEETKDILL 170
Query: 404 LDVMPLSLGIEVVEGVMSVLIPKNTTIPTKKERTFRTTSDNQTSVWIKVYEGEGAKTDKN 463
LDV PL+LGIE V GVM+ LIP+NT IPTKK + F T D QT+V I+V+EGE + T
Sbjct: 171 LDVAPLTLGIETVGGVMTKLIPRNTVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDC 350
Query: 464 FFLGKFELSGFSPAPKGDTEINVCFDVDADGIVEVSAED 502
LGKF+LSG PAP+G +I V F+VDA+GI+ V AED
Sbjct: 351 RLLGKFDLSGIPPAPRGTPQIEVTFEVDANGILNVKAED 467
>BP064075
Length = 554
Score = 178 bits (451), Expect = 3e-45
Identities = 91/159 (57%), Positives = 120/159 (75%)
Frame = -2
Query: 287 TIELDSLCGGIDLHVTVTQGLFEKLNKDLFEKCMEIVEKCLVEAKIGKSEVHELVLVGGS 346
TIELD L GID + ++T+ FE+LNKD F+KCME+VEKC++++K+ KS +H++VLVGGS
Sbjct: 553 TIELDFLYQGIDFYSSITRAKFEELNKDYFQKCMELVEKCVIDSKMDKSNIHDVVLVGGS 374
Query: 347 TRIPKVQQLLKEMFSVHGRVKELCKSINPDEAVAYGAAVQAAILSSEGDKKVEELLLLDV 406
+RIPKV+QLL + F GR K+LC INPDEAVA+GAAV A+ILS E +KV+ LLL +V
Sbjct: 373 SRIPKVRQLLMDFF---GR-KDLCNRINPDEAVAHGAAVHASILSGEFSEKVQSLLLREV 206
Query: 407 MPLSLGIEVVEGVMSVLIPKNTTIPTKKERTFRTTSDNQ 445
PLSLG+E G+M +IP+NT IPT + F T NQ
Sbjct: 205 TPLSLGLEKHGGIMETIIPRNTMIPTTMDHVFTTHFHNQ 89
>AV766006
Length = 408
Score = 139 bits (351), Expect = 1e-33
Identities = 73/136 (53%), Positives = 97/136 (70%)
Frame = -1
Query: 416 VEGVMSVLIPKNTTIPTKKERTFRTTSDNQTSVWIKVYEGEGAKTDKNFFLGKFELSGFS 475
V GVMS +I +NT IPT+K +TF T D QT+V I+VYEGE A T N LGKF+L+G
Sbjct: 408 VGGVMSKIIERNTVIPTRKTQTFTTYQDKQTTVSIQVYEGERAMTKDNRQLGKFDLNGIP 229
Query: 476 PAPKGDTEINVCFDVDADGIVEVSAEDMSLRLKKTITITNRLGRLSQEEMRRMVRDAAQY 535
A +G +I V F+VDA+GI+ VSAED + K+ ITITN GRLS+EE+ RMV++A ++
Sbjct: 228 AAARGTPQIEVTFEVDANGILAVSAEDKASSAKEAITITNDKGRLSKEEIERMVKEAEEF 49
Query: 536 KAEDEEVRKKVKAKNS 551
D EV++KV+ KNS
Sbjct: 48 ADADAEVKRKVEGKNS 1
>AW163930
Length = 359
Score = 137 bits (345), Expect = 5e-33
Identities = 69/89 (77%), Positives = 78/89 (87%)
Frame = +1
Query: 367 KELCKSINPDEAVAYGAAVQAAILSSEGDKKVEELLLLDVMPLSLGIEVVEGVMSVLIPK 426
KELCKSINPDEAVAYGAAVQAAILS EG++KV++LLLLDV PLSLG+E GVM+VLIP+
Sbjct: 52 KELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPR 231
Query: 427 NTTIPTKKERTFRTTSDNQTSVWIKVYEG 455
NTTIPTKKE+ F T SDNQ V I+VYEG
Sbjct: 232 NTTIPTKKEQVFSTYSDNQPGVLIQVYEG 318
>AV769455
Length = 556
Score = 117 bits (292), Expect = 7e-27
Identities = 65/184 (35%), Positives = 99/184 (53%), Gaps = 1/184 (0%)
Frame = +1
Query: 54 MGDAAINQLALNPHNTVFDAKRLIGRRFSDRSVQQDMKLWPFKVVQGTRDKPMISVTYKG 113
+G A + +NP N++ KRLIGR+F+D +Q+D+K PF V +G P+I Y G
Sbjct: 4 LGTAGASSTMMNPKNSISQLKRLIGRQFADPELQRDLKSLPFAVTEGPDGFPLIHARYLG 183
Query: 114 EERKLAAEEISSMVLFKMKEVAEAYLGHEVKRAVITVPAYFNNAQRQSTKDAGEIAGFDV 173
E R ++ M+L +KE+A+ L V I +P YF + QR++ DA IAG
Sbjct: 184 ETRTFTPTQVFGMMLSNLKEIAQKNLNAAVVDCCIGIPIYFTDLQRRAVLDAATIAGLHP 363
Query: 174 MRIINEPTAASIAYGLDKKGWRKGEQ-NVLVFDLGGGTFDVSLVTIDEGIFKVNATVGDT 232
+R+ +E TA ++AYG+ K + + NV D+G + V + +G KV A D
Sbjct: 364 LRLFHETTATALAYGIYKTDLPENDPLNVAFVDVGHASMQVCIAGYKKGQLKVLAHSYDR 543
Query: 233 YLGG 236
LGG
Sbjct: 544 SLGG 555
>AV412686
Length = 166
Score = 113 bits (282), Expect = 1e-25
Identities = 55/55 (100%), Positives = 55/55 (100%)
Frame = +2
Query: 230 GDTYLGGVDFDNNLVNFLVDFFQRKHNKDITENVKSLRRLRLACEKAKRILSSTS 284
GDTYLGGVDFDNNLVNFLVDFFQRKHNKDITENVKSLRRLRLACEKAKRILSSTS
Sbjct: 2 GDTYLGGVDFDNNLVNFLVDFFQRKHNKDITENVKSLRRLRLACEKAKRILSSTS 166
>BP083571
Length = 423
Score = 101 bits (251), Expect = 4e-22
Identities = 52/72 (72%), Positives = 58/72 (80%), Gaps = 2/72 (2%)
Frame = +1
Query: 1 MATSKKDKAIGIDLGTTYSCVAVWN--HNRVEIIPNELGNRTTPSYVAFTDTERLMGDAA 58
MAT + AIGIDLGTTYSCVAVW ++R EII NE GNRTTPS+VAFTDT+RL+GDAA
Sbjct: 145 MATKYEGPAIGIDLGTTYSCVAVWQEQNDRAEIIHNEQGNRTTPSFVAFTDTQRLIGDAA 324
Query: 59 INQLALNPHNTV 70
NQ A NP NTV
Sbjct: 325 KNQAATNPTNTV 360
>CB827219
Length = 544
Score = 90.1 bits (222), Expect = 9e-19
Identities = 49/163 (30%), Positives = 94/163 (57%)
Frame = +3
Query: 194 WRKGEQNVLVFDLGGGTFDVSLVTIDEGIFKVNATVGDTYLGGVDFDNNLVNFLVDFFQR 253
W G + +L F++G G DV++ G+ ++ A G T +GG D N+++ L+ +
Sbjct: 21 WAVGVRKLLYFNMGAGYCDVAVTVTAGGVSQIKALAGST-IGGEDLLQNMMHHLLPDCEN 197
Query: 254 KHNKDITENVKSLRRLRLACEKAKRILSSTSQTTIELDSLCGGIDLHVTVTQGLFEKLNK 313
+ +KS+ LR+A A LSS +++D L G+ ++ + FE++NK
Sbjct: 198 IFKNHGVKEIKSMGLLRVATLDAIHRLSSQKSVQVDVD-LGDGLKINKVIDCEEFEEVNK 374
Query: 314 DLFEKCMEIVEKCLVEAKIGKSEVHELVLVGGSTRIPKVQQLL 356
++FEKC ++ +CL ++KI +V+++++VGG + IP+V+ L+
Sbjct: 375 NVFEKCESLIIQCLQDSKIEVEDVNDVIIVGGCSYIPRVKNLV 503
>BP033771
Length = 513
Score = 88.2 bits (217), Expect = 3e-18
Identities = 38/85 (44%), Positives = 65/85 (75%)
Frame = -2
Query: 484 INVCFDVDADGIVEVSAEDMSLRLKKTITITNRLGRLSQEEMRRMVRDAAQYKAEDEEVR 543
+NVCFD+DA+GI+ VSAE+ + +K IT+TN GRLS E++ R++++A +++AED
Sbjct: 509 LNVCFDLDANGILNVSAEEETTGIKNGITVTNDNGRLSSEQIMRLIQEAEKHRAEDRMYE 330
Query: 544 KKVKAKNSLENYAYEARDRVKKLEK 568
KKV+A N+L++Y Y+ R+ +K +++
Sbjct: 329 KKVEAMNALDDYVYKLRNAIKDIDR 255
>AV409958
Length = 389
Score = 81.6 bits (200), Expect = 3e-16
Identities = 47/121 (38%), Positives = 74/121 (60%), Gaps = 4/121 (3%)
Frame = +2
Query: 243 LVNFLVDFFQRKHNKDITENVKSLRRLRLACEKAKRILSSTSQTTIELDSLCGGID---- 298
+V++L F+R D+ ++ ++L+RL EKAK LSS +QT I L + D
Sbjct: 5 IVDWLAADFKRNEGIDLLKDKQALQRLTETAEKAKMELSSLTQTNISLPFITATADGPKH 184
Query: 299 LHVTVTQGLFEKLNKDLFEKCMEIVEKCLVEAKIGKSEVHELVLVGGSTRIPKVQQLLKE 358
+ T+T+ FE+L DL ++ VE L +AK+ ++ E++LVGGSTRIP VQ+L+K+
Sbjct: 185 IETTLTRAKFEELCSDLLDRLRTPVENSLRDAKLSFKDLDEVILVGGSTRIPAVQELVKK 364
Query: 359 M 359
M
Sbjct: 365 M 367
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.134 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,076,569
Number of Sequences: 28460
Number of extensions: 91183
Number of successful extensions: 427
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 407
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 411
length of query: 602
length of database: 4,897,600
effective HSP length: 95
effective length of query: 507
effective length of database: 2,193,900
effective search space: 1112307300
effective search space used: 1112307300
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0118c.15


