

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0118c.11
(259 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
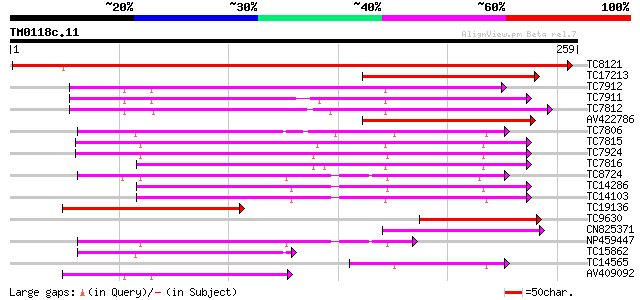
Score E
Sequences producing significant alignments: (bits) Value
TC8121 UP|Q9LKJ5 (Q9LKJ5) Multifunctional transport intrinsic me... 383 e-107
TC17213 similar to UP|AAS48064 (AAS48064) NIP2, partial (30%) 146 4e-36
TC7912 similar to UP|Q39800 (Q39800) Delta-tonoplast intrinsic p... 117 2e-27
TC7911 similar to UP|Q39800 (Q39800) Delta-tonoplast intrinsic p... 114 1e-26
TC7812 UP|Q9LKJ6 (Q9LKJ6) Water-selective transport intrinsic me... 108 1e-24
AV422786 106 5e-24
TC7806 homologue to UP|Q946J9 (Q946J9) Aquaporin protein PIP1,1,... 98 1e-21
TC7815 similar to UP|Q9AVB5 (Q9AVB5) Plasma membrane intrinsic p... 93 5e-20
TC7924 homologue to UP|CAE53883 (CAE53883) Aquaporin, partial (95%) 92 7e-20
TC7816 homologue to UP|Q9ATM4 (Q9ATM4) Plasma membrane integral ... 92 9e-20
TC8724 homologue to UP|Q9AQU5 (Q9AQU5) Plasma membrane integral ... 91 2e-19
TC14286 homologue to UP|O65357 (O65357) Aquaporin 2, complete 91 2e-19
TC14103 similar to UP|Q84XC6 (Q84XC6) Plasma intrinsic protein 2... 85 1e-17
TC19136 similar to UP|AAQ11827 (AAQ11827) Nodulin-like intrinsic... 84 2e-17
TC9630 similar to UP|AAS48063 (AAS48063) NIP3, partial (18%) 70 3e-13
CN825371 61 2e-10
NP459447 plasma membrane intrinsic protein homolog [Lotus japoni... 54 3e-08
TC15862 homologue to UP|O22339 (O22339) Aquaporin-like transmemb... 53 5e-08
TC14565 homologue to UP|O65741 (O65741) Transmembrane channel pr... 49 7e-07
AV409092 48 2e-06
>TC8121 UP|Q9LKJ5 (Q9LKJ5) Multifunctional transport intrinsic membrane
protein 2, complete
Length = 1213
Score = 383 bits (983), Expect = e-107
Identities = 187/257 (72%), Positives = 222/257 (85%), Gaps = 1/257 (0%)
Frame = +2
Query: 2 ADEVVLNVNNEASKKCDSIEED-CVPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVT 60
A EVV+NVN +ASK + + + V LQK++AE++GTYF IFAGCAS+VVN NND VVT
Sbjct: 128 AHEVVVNVNKDASKTIEVSDTNFTVSFLQKVIAELVGTYFFIFAGCASIVVNKNNDNVVT 307
Query: 61 LPGIAIVWGLAVMVLVYSIGHISGAHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLA 120
LPGIA+VWGLAVMVLVYS+GHISGAHFNPA TIA +TKRFP KQVPAY+ AQV+GSTLA
Sbjct: 308 LPGIALVWGLAVMVLVYSLGHISGAHFNPAATIAFASTKRFPWKQVPAYVSAQVLGSTLA 487
Query: 121 SGTLRLIFSGKDNHFTGTLPAGSDLQAFVVEFIITFLLMFVVSGVATDNRAIGELAGLAV 180
SGTLRLIFSGK N F G LP GS+LQAFV+EFIITF L+F++ GVATD+RAIGE+AG+ V
Sbjct: 488 SGTLRLIFSGKHNQFAGALPTGSNLQAFVIEFIITFFLIFILFGVATDDRAIGEVAGIVV 667
Query: 181 GSTVMLNVLFAGPITGASMNPARSLGPAIVHNQYKGIWIYMVSPILGAVAGTWAYSFIRI 240
GSTV+LNVLFAGPITGASMNPARS+G A VHN+Y+GIWIY++SP LGAVAG W Y+ +R
Sbjct: 668 GSTVLLNVLFAGPITGASMNPARSIGSAFVHNEYRGIWIYLLSPTLGAVAGAWVYNIVRY 847
Query: 241 TNKPVRELTKSSSFLKG 257
T+KP+RE+TK+ SFLKG
Sbjct: 848 TDKPLREITKNVSFLKG 898
>TC17213 similar to UP|AAS48064 (AAS48064) NIP2, partial (30%)
Length = 578
Score = 146 bits (368), Expect = 4e-36
Identities = 65/81 (80%), Positives = 79/81 (97%)
Frame = +3
Query: 162 VSGVATDNRAIGELAGLAVGSTVMLNVLFAGPITGASMNPARSLGPAIVHNQYKGIWIYM 221
+SGV+TDNRAIGE+AGLA+G+T++LNV+ AGPITGASMNPARSLGPAIVH++YKGIWIY+
Sbjct: 3 ISGVSTDNRAIGEMAGLAIGATILLNVIIAGPITGASMNPARSLGPAIVHSEYKGIWIYL 182
Query: 222 VSPILGAVAGTWAYSFIRITN 242
VSP+LGAVAGTW+Y+FIR TN
Sbjct: 183 VSPVLGAVAGTWSYNFIRYTN 245
>TC7912 similar to UP|Q39800 (Q39800) Delta-tonoplast intrinsic protein,
partial (95%)
Length = 1045
Score = 117 bits (294), Expect = 2e-27
Identities = 71/207 (34%), Positives = 106/207 (50%), Gaps = 7/207 (3%)
Frame = +2
Query: 28 LQKLVAEVLGTYFLIFAGCASVVV--NLNNDKVVTLPG---IAIVWGLAVMVLVYSIGHI 82
++ +AE + T +FAG S + + +D + G IA+ A+ V V +I
Sbjct: 80 IKAYIAEFISTLLFVFAGVGSAIAYGKVTSDAALDPAGLVAIAVCHAFALFVAVSVGANI 259
Query: 83 SGAHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGKDNHFTGTLPAG 142
SG H NPAVT + + Y +AQ++GS +A L + G + G
Sbjct: 260 SGGHVNPAVTFGLALGGQITILTGIFYWIAQLLGSIVACFLLHYVTGGLETPVHGLAAGV 439
Query: 143 SDLQAFVVEFIITFLLMFVVSGVATDNR--AIGELAGLAVGSTVMLNVLFAGPITGASMN 200
+ V E IITF L++ V A D + ++G +A +A+G V N+L AGP +G SMN
Sbjct: 440 GAFEGVVTEIIITFGLVYTVYATAADPKKGSLGTIAPIAIGFIVGANILAAGPFSGGSMN 619
Query: 201 PARSLGPAIVHNQYKGIWIYMVSPILG 227
PARS GPA+V + IWIY V P++G
Sbjct: 620 PARSFGPAVVSGNFHDIWIYWVGPLIG 700
>TC7911 similar to UP|Q39800 (Q39800) Delta-tonoplast intrinsic protein,
complete
Length = 1018
Score = 114 bits (286), Expect = 1e-26
Identities = 76/222 (34%), Positives = 112/222 (50%), Gaps = 11/222 (4%)
Frame = +1
Query: 28 LQKLVAEVLGTYFLIFAGCASVVV--NLNNDKVVTLPG---IAIVWGLAVMVLVYSIGHI 82
++ +AE + T +FAG S + L +D + G +AI A+ V V +I
Sbjct: 109 IKAYIAEFISTLIFVFAGVGSAIAYGKLTSDAALDPAGLVAVAICHAFALFVAVSVGANI 288
Query: 83 SGAHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGKDNHFTGTLP-- 140
SG H NPAVT + L Y +AQ++GS +A L+ + G TLP
Sbjct: 289 SGGHVNPAVTFGLALGGQITLLTGLFYWIAQLLGSIVACFLLKFVTGGL------TLPIH 450
Query: 141 --AGSDLQAFVVEFIITFLLMFVVSGVATDNR--AIGELAGLAVGSTVMLNVLFAGPITG 196
A + V E IITF L++ V A D + ++G +A +A+G V N+L AGP +G
Sbjct: 451 SVAAGAGEGVVTEIIITFGLVYTVYATAADPKKGSLGTIAPIAIGFIVGANILAAGPFSG 630
Query: 197 ASMNPARSLGPAIVHNQYKGIWIYMVSPILGAVAGTWAYSFI 238
SMNPARS GPA+V + WIY V P++G Y+++
Sbjct: 631 GSMNPARSFGPAVVSGDFHDNWIYWVGPLIGGGLAGVIYTYV 756
>TC7812 UP|Q9LKJ6 (Q9LKJ6) Water-selective transport intrinsic membrane
protein 1, complete
Length = 1149
Score = 108 bits (269), Expect = 1e-24
Identities = 76/229 (33%), Positives = 114/229 (49%), Gaps = 8/229 (3%)
Frame = +3
Query: 28 LQKLVAEVLGTYFLIFAGCASVVV--NLNNDKVVTLPGI---AIVWGLAVMVLVYSIGHI 82
L+ +AE + T+ +FAG S + L +D T G+ +I A+ V V +I
Sbjct: 132 LRSALAEFISTFIFVFAGSGSGIAYNKLTDDGAATPAGLISASIAHAFALFVAVSISANI 311
Query: 83 SGAHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGKDNHFTGTLPAG 142
SG H NPAVT + YI+AQ++GS +AS L + F L AG
Sbjct: 312 SGGHVNPAVTFGLFVGGNISFLRGVIYIIAQLLGSIVASLLLAFVTGLAVPAFG--LSAG 485
Query: 143 SDL-QAFVVEFIITFLLMFVVSGVATDNR--AIGELAGLAVGSTVMLNVLFAGPITGASM 199
+ A V+E ++TF L++ V A D + ++G +A +A+G V N+L G GASM
Sbjct: 486 VGVGNALVLEIVMTFGLVYTVYATAVDPKKGSLGTIAPIAIGFIVGANILLGGAFDGASM 665
Query: 200 NPARSLGPAIVHNQYKGIWIYMVSPILGAVAGTWAYSFIRITNKPVREL 248
NPA S GPA+V + WIY V P++G Y + I ++ +L
Sbjct: 666 NPAVSFGPAVVSWSWSNHWIYWVGPLVGGGIAGLIYEVVFIGSQTHEQL 812
>AV422786
Length = 427
Score = 106 bits (264), Expect = 5e-24
Identities = 46/79 (58%), Positives = 64/79 (80%)
Frame = -3
Query: 162 VSGVATDNRAIGELAGLAVGSTVMLNVLFAGPITGASMNPARSLGPAIVHNQYKGIWIYM 221
V+ VATD+RA+GELAG+AVG+TV+LN+L +GP +G SMNP R+LGPA+ YK IWIY+
Sbjct: 425 VTAVATDSRAVGELAGIAVGATVLLNILISGPTSGGSMNPVRTLGPAVAAGNYKHIWIYL 246
Query: 222 VSPILGAVAGTWAYSFIRI 240
V+P LGA+AG Y+ +++
Sbjct: 245 VAPTLGALAGAGVYTLVKL 189
>TC7806 homologue to UP|Q946J9 (Q946J9) Aquaporin protein PIP1,1, complete
Length = 1413
Score = 98.2 bits (243), Expect = 1e-21
Identities = 71/216 (32%), Positives = 107/216 (48%), Gaps = 19/216 (8%)
Frame = +3
Query: 32 VAEVLGTYFLIFAGCASVVVNLNND---KVVTLPGIAIVWGLAVMVLVYSIGHISGAHFN 88
+AE + T+ ++ +V+ D K V + GIA +G + LVY ISG H N
Sbjct: 240 IAEFVATFLFLYITVLTVMGVSGADSKCKTVGIQGIAWAFGGMIFALVYCTAGISGGHIN 419
Query: 89 PAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGKDNHFTGTLPAGSDLQA- 147
PAVT ++ L + YI+ Q +G+ + +G ++ F GK + GTL G++ A
Sbjct: 420 PAVTFGLFLARKLSLTRAIFYIVMQCLGAIVGAGVVK-GFEGKTKY--GTLGGGANFVAP 590
Query: 148 -------FVVEFIITFLLMFVVSGVATDNRAIGE-----LAGLAVGSTVMLNVLFAGPIT 195
E + TF+L++ V R+ + LA L +G V L L PIT
Sbjct: 591 GYTKGDGLGAEIVGTFVLVYTVFSATDAKRSARDSHVPILAPLPIGFAVFLVHLATIPIT 770
Query: 196 GASMNPARSLGPAIVHNQYKG---IWIYMVSPILGA 228
G +NPARSLG AI+ N+ +G WI+ V P +GA
Sbjct: 771 GTGINPARSLGAAIIFNKDRGWDDHWIFWVGPFIGA 878
Score = 33.9 bits (76), Expect = 0.029
Identities = 28/103 (27%), Positives = 47/103 (45%), Gaps = 10/103 (9%)
Frame = +3
Query: 139 LPAGSDLQAFVVEFIITFLLMFV-------VSGVATDNRAIGELAGL--AVGSTVMLNVL 189
L + S +A + EF+ TFL +++ VSG + + +G + G+ A G + V
Sbjct: 210 LTSWSFYRAGIAEFVATFLFLYITVLTVMGVSGADSKCKTVG-IQGIAWAFGGMIFALVY 386
Query: 190 FAGPITGASMNPARSLGPAIVHN-QYKGIWIYMVSPILGAVAG 231
I+G +NPA + G + Y+V LGA+ G
Sbjct: 387 CTAGISGGHINPAVTFGLFLARKLSLTRAIFYIVMQCLGAIVG 515
>TC7815 similar to UP|Q9AVB5 (Q9AVB5) Plasma membrane intrinsic protein
2-1, complete
Length = 1450
Score = 92.8 bits (229), Expect = 5e-20
Identities = 66/225 (29%), Positives = 104/225 (45%), Gaps = 17/225 (7%)
Frame = +1
Query: 31 LVAEVLGTYFLIFAGCASVVVNLNNDKV-----VTLPGIAIVWGLAVMVLVYSIGHISGA 85
L+AE + T ++ +V+ + + + GIA +G + +LVY ISG
Sbjct: 184 LIAEFIATLLFLYVTVLTVIGHSRLNSADPCDGAGILGIAWAFGGMIFILVYCTAGISGG 363
Query: 86 HFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGKDNHFTGTL----PA 141
H NPAVT ++ L + YI+AQ +G+ G + N + G P
Sbjct: 364 HINPAVTFGLFIGRKVSLIRALLYIVAQCLGAISGVGLAKAFQKSFYNRYGGAANFVQPG 543
Query: 142 GSDLQAFVVEFIITFLLMFVVSGVATDNR-----AIGELAGLAVGSTVMLNVLFAGPITG 196
S+ A E I TF+L++ V R + LA L +G V + L P+TG
Sbjct: 544 YSNGTALGAEIIGTFVLVYTVFSATDPKRNARDSHVPVLAPLPIGFAVFMVHLATIPVTG 723
Query: 197 ASMNPARSLGPAIVHNQYK---GIWIYMVSPILGAVAGTWAYSFI 238
+NPARS G A+++N+ K WI+ V P +GA+A + +I
Sbjct: 724 TGINPARSFGAAVIYNEDKIWDDHWIFWVGPFIGALAAAIYHQYI 858
>TC7924 homologue to UP|CAE53883 (CAE53883) Aquaporin, partial (95%)
Length = 1078
Score = 92.4 bits (228), Expect = 7e-20
Identities = 69/223 (30%), Positives = 102/223 (44%), Gaps = 15/223 (6%)
Frame = +2
Query: 31 LVAEVLGTYFLIFAGCASVVVNLNNDKV---VTLPGIAIVWGLAVMVLVYSIGHISGAHF 87
L+AE + T ++ A+V+ + V L GIA +G + VLVY ISG H
Sbjct: 131 LIAEFIATLLFLYVTVATVIGHKKQTGPCDGVGLLGIAWSFGGMIFVLVYCTAGISGGHI 310
Query: 88 NPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGKDNHFTG----TLPAGS 143
NPAVT ++ L + Y++AQ +G+ G ++ N G S
Sbjct: 311 NPAVTFGLFLARKVSLIRAVLYMVAQCLGAICGVGLVKAFMKHPYNSLGGGANSVAHGYS 490
Query: 144 DLQAFVVEFIITFLLMFVVSGVATDNRA-----IGELAGLAVGSTVMLNVLFAGPITGAS 198
A E I TF+L++ V R+ + LA L +G V + L PITG
Sbjct: 491 KGSALGAEIIGTFVLVYTVFSATDPKRSARDSHVPVLAPLPIGFAVFMVHLATIPITGTG 670
Query: 199 MNPARSLGPAIVHNQYK---GIWIYMVSPILGAVAGTWAYSFI 238
+NPARS G A++ N K WI+ V P +GA+A + +I
Sbjct: 671 INPARSFGAAVIFNNGKVWDDQWIFWVGPFVGALAAAAYHQYI 799
>TC7816 homologue to UP|Q9ATM4 (Q9ATM4) Plasma membrane integral protein
ZmPIP2-7, complete
Length = 1523
Score = 92.0 bits (227), Expect = 9e-20
Identities = 64/192 (33%), Positives = 93/192 (48%), Gaps = 12/192 (6%)
Frame = +1
Query: 59 VTLPGIAIVWGLAVMVLVYSIGHISGAHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGST 118
V + GIA +G + VLVY ISG H NPAVT ++ L + YI+AQ +G+
Sbjct: 409 VGILGIAWAFGGMIFVLVYCTAGISGGHINPAVTFGLFVGRKVSLVRALLYIIAQCLGAI 588
Query: 119 LASGTLRLIFSGKDNHFTG---TLPAG-SDLQAFVVEFIITFLLMFVVSGVATDNR---- 170
+G + N + G T+ G S A E I TF+L++ V R
Sbjct: 589 CGAGLAKGFQKSFYNRYGGGANTISDGYSKGTALGAEIIGTFVLVYTVFSATDPKRNARD 768
Query: 171 -AIGELAGLAVGSTVMLNVLFAGPITGASMNPARSLGPAIVHNQYK---GIWIYMVSPIL 226
+ LA L +G V + L P+TG +NPARS GPA++ N+ K W+Y V P +
Sbjct: 769 SHVPVLAPLPIGFAVFMVHLATIPVTGTGINPARSFGPAVIFNEDKVWDDQWVYWVGPFV 948
Query: 227 GAVAGTWAYSFI 238
GA+ + +I
Sbjct: 949 GALVAAIYHQYI 984
>TC8724 homologue to UP|Q9AQU5 (Q9AQU5) Plasma membrane integral protein
ZmPIP1-4 (Plasma membrane integral protein ZmPIP1-3),
partial (93%)
Length = 1560
Score = 91.3 bits (225), Expect = 2e-19
Identities = 71/217 (32%), Positives = 107/217 (48%), Gaps = 20/217 (9%)
Frame = +1
Query: 32 VAEVLGTYFLIFAGCASVV-VNLNNDKV--VTLPGIAIVWGLAVMVLVYSIGHISGAHFN 88
+AE + T+ ++ +V+ VN + +K V + GIA +G + LVY ISG H N
Sbjct: 250 IAEFVATFLFLYITVLTVMGVNRSPNKCSSVGIQGIAWAFGGMIFALVYCTAGISGGHIN 429
Query: 89 PAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLR--------LIFSGKDNHFTGTLP 140
PAVT ++ L + YI+ Q +G+ +G ++ +F G N
Sbjct: 430 PAVTFGLFLARKLSLTRALFYIVMQCLGAICGAGVVKGFEGNARFEMFKGGANVVNPGYT 609
Query: 141 AGSDLQAFVVEFIITFLLMFVVSGVATDNRA------IGELAGLAVGSTVMLNVLFAGPI 194
G L A E + TF+L++ V ATD + + LA L +G V L L PI
Sbjct: 610 KGDGLGA---EIVGTFVLVYTVFS-ATDAKRNARDSHVPLLAPLPIGFAVFLVHLATIPI 777
Query: 195 TGASMNPARSLGPAIVHNQ---YKGIWIYMVSPILGA 228
TG +NPARSLG AI++N+ + WI+ V P +GA
Sbjct: 778 TGTGINPARSLGAAIIYNRDHAWDDHWIFWVGPFIGA 888
Score = 33.5 bits (75), Expect = 0.037
Identities = 28/104 (26%), Positives = 46/104 (43%), Gaps = 9/104 (8%)
Frame = +1
Query: 137 GTLPAGSDLQAFVVEFIITFLLMFV-VSGVATDNRAIGELAGL-------AVGSTVMLNV 188
G L + S +A + EF+ TFL +++ V V NR+ + + + A G + V
Sbjct: 214 GELKSWSFYRAGIAEFVATFLFLYITVLTVMGVNRSPNKCSSVGIQGIAWAFGGMIFALV 393
Query: 189 LFAGPITGASMNPARSLGPAIVHN-QYKGIWIYMVSPILGAVAG 231
I+G +NPA + G + Y+V LGA+ G
Sbjct: 394 YCTAGISGGHINPAVTFGLFLARKLSLTRALFYIVMQCLGAICG 525
>TC14286 homologue to UP|O65357 (O65357) Aquaporin 2, complete
Length = 1278
Score = 90.9 bits (224), Expect = 2e-19
Identities = 62/195 (31%), Positives = 92/195 (46%), Gaps = 15/195 (7%)
Frame = +1
Query: 59 VTLPGIAIVWGLAVMVLVYSIGHISGAHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGST 118
V + GIA +G + +LVY ISG H NPAVT ++ L + Y++AQ +G+
Sbjct: 337 VGILGIAWAFGGMIFILVYCTAGISGGHINPAVTFGLFLARKVSLIRAILYMVAQCLGAI 516
Query: 119 LASGTLRLI-------FSGKDNHFTGTLPAGSDLQAFVVEFIITFLLMFVVSGVATDNRA 171
G ++ + G N G+ L A E I TF+L++ V R+
Sbjct: 517 CGVGLVKAFQKSHYNKYGGGANSLNDGYSTGTGLGA---EIIGTFVLVYTVFSATDPKRS 687
Query: 172 -----IGELAGLAVGSTVMLNVLFAGPITGASMNPARSLGPAIVHNQYK---GIWIYMVS 223
+ LA L +G V + L P+TG +NPARS G A++ NQ K WI+ V
Sbjct: 688 ARDSHVPVLAPLPIGFAVFMVHLATIPVTGTGINPARSFGAAVIFNQSKPWDDHWIFWVG 867
Query: 224 PILGAVAGTWAYSFI 238
P +GA + + FI
Sbjct: 868 PFIGAAIAAFYHQFI 912
>TC14103 similar to UP|Q84XC6 (Q84XC6) Plasma intrinsic protein 2,2, partial
(94%)
Length = 1199
Score = 85.1 bits (209), Expect = 1e-17
Identities = 60/195 (30%), Positives = 90/195 (45%), Gaps = 15/195 (7%)
Frame = +3
Query: 59 VTLPGIAIVWGLAVMVLVYSIGHISGAHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGST 118
V + GIA +G + +LVY ISG H NPAVT ++ L + Y++AQ +G+
Sbjct: 321 VGILGIAWAFGGMIFILVYCTAGISGGHINPAVTFGLFLARKVSLVRALFYMVAQCLGAI 500
Query: 119 LASGTLRLI-------FSGKDNHFTGTLPAGSDLQAFVVEFIITFLLMFVVSGVATDNR- 170
G ++ + G N G+ L A E I TF+L++ V R
Sbjct: 501 SGVGLVKAFQKSYYNRYHGGANMLADGYSKGTGLGA---EIIGTFVLVYTVFSATDPKRN 671
Query: 171 ----AIGELAGLAVGSTVMLNVLFAGPITGASMNPARSLGPAIVHNQYKG---IWIYMVS 223
+ LA L +G V + L PITG +NPARS G A+++N K WI+ V
Sbjct: 672 ARDSHVPVLAPLPIGFAVFMVHLATIPITGTGINPARSFGAAVIYNNEKAWDDQWIFWVG 851
Query: 224 PILGAVAGTWAYSFI 238
P +GA + ++
Sbjct: 852 PFIGATIAAIYHQYV 896
>TC19136 similar to UP|AAQ11827 (AAQ11827) Nodulin-like intrinsic protein
NIP1-2 (Fragment), partial (29%)
Length = 556
Score = 84.0 bits (206), Expect = 2e-17
Identities = 43/83 (51%), Positives = 53/83 (63%)
Frame = +2
Query: 25 VPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISG 84
VPL QK++AE +GT+ LIFA A +VN D L G A GL VM ++ SIGHISG
Sbjct: 308 VPLTQKILAEFVGTFILIFAATAGPIVNNKYDGAEGLMGNAATAGLTVMFIILSIGHISG 487
Query: 85 AHFNPAVTIAHVTTKRFPLKQVP 107
AH NP++TIA + FP QVP
Sbjct: 488 AHLNPSLTIAFAAFRHFPWSQVP 556
Score = 35.4 bits (80), Expect = 0.010
Identities = 22/72 (30%), Positives = 35/72 (48%), Gaps = 5/72 (6%)
Frame = +2
Query: 139 LPAGSDLQAFVVEFIITFLLMFVVSGVATDNRAIGELAGL-----AVGSTVMLNVLFAGP 193
+P+ Q + EF+ TF+L+F + N GL G TVM +L G
Sbjct: 299 IPSVPLTQKILAEFVGTFILIFAATAGPIVNNKYDGAEGLMGNAATAGLTVMFIILSIGH 478
Query: 194 ITGASMNPARSL 205
I+GA +NP+ ++
Sbjct: 479 ISGAHLNPSLTI 514
>TC9630 similar to UP|AAS48063 (AAS48063) NIP3, partial (18%)
Length = 971
Score = 70.5 bits (171), Expect = 3e-13
Identities = 28/56 (50%), Positives = 43/56 (76%)
Frame = +3
Query: 188 VLFAGPITGASMNPARSLGPAIVHNQYKGIWIYMVSPILGAVAGTWAYSFIRITNK 243
+L+ P TGASMNP R+LGPAI N YK IW+Y+V+P+LGA++G Y+ +++ ++
Sbjct: 177 ILYCRPETGASMNPVRTLGPAIAANNYKAIWVYLVAPVLGALSGAGIYTAVKLPDE 344
>CN825371
Length = 483
Score = 60.8 bits (146), Expect = 2e-10
Identities = 28/74 (37%), Positives = 43/74 (57%)
Frame = +1
Query: 171 AIGELAGLAVGSTVMLNVLFAGPITGASMNPARSLGPAIVHNQYKGIWIYMVSPILGAVA 230
+IG +A LA+G V N+L GP GA MNPA + GP++V ++ W++ + P LGA
Sbjct: 13 SIGAIAPLAIGLIVGANILVGGPFDGACMNPALAFGPSLVGWRWHYHWVFWLGPFLGAAL 192
Query: 231 GTWAYSFIRITNKP 244
Y ++ I +P
Sbjct: 193 AALIYEYLVIPTEP 234
>NP459447 plasma membrane intrinsic protein homolog [Lotus japonicus]
Length = 578
Score = 53.5 bits (127), Expect = 3e-08
Identities = 48/171 (28%), Positives = 75/171 (43%), Gaps = 16/171 (9%)
Frame = +1
Query: 32 VAEVLGTYFLIFAGCASVVVNLNNDKV---VTLPGIAIVWGLAVMVLVYSIGHISGAHFN 88
+AE + T+ ++ +V+ + + V + GIA +G + LVY ISG H N
Sbjct: 73 IAEFIATFLFLYITVLTVMGVKRSPNMCASVGIQGIAWAFGGMIFALVYCTAGISGGHIN 252
Query: 89 PAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLR-------LIFSGKDNHFTGTLPA 141
PAVT ++ L + YI+ Q +G+ +G ++ G N
Sbjct: 253 PAVTFGLFLARKLSLTRAVYYIVMQCLGAICGAGVVKGFQPKQYQALGGGANTIAHGYTK 432
Query: 142 GSDLQAFVVEFIITFLLMFVVSGVATDNRA------IGELAGLAVGSTVML 186
GS L A E I TF+L++ V ATD + + LA L +G V L
Sbjct: 433 GSGLGA---EIIGTFVLVYTVFS-ATDAKRNARDSHVPILAPLPIGFAVFL 573
Score = 31.6 bits (70), Expect = 0.14
Identities = 28/104 (26%), Positives = 42/104 (39%), Gaps = 9/104 (8%)
Frame = +1
Query: 137 GTLPAGSDLQAFVVEFIITFLLMFV----VSGVATDNRAIGELA----GLAVGSTVMLNV 188
G L + S +A + EFI TFL +++ V GV + A G + V
Sbjct: 37 GELASWSFWRAGIAEFIATFLFLYITVLTVMGVKRSPNMCASVGIQGIAWAFGGMIFALV 216
Query: 189 LFAGPITGASMNPARSLGPAIVHN-QYKGIWIYMVSPILGAVAG 231
I+G +NPA + G + Y+V LGA+ G
Sbjct: 217 YCTAGISGGHINPAVTFGLFLARKLSLTRAVYYIVMQCLGAICG 348
>TC15862 homologue to UP|O22339 (O22339) Aquaporin-like transmembrane
channel protein, partial (61%)
Length = 576
Score = 53.1 bits (126), Expect = 5e-08
Identities = 32/103 (31%), Positives = 53/103 (51%), Gaps = 3/103 (2%)
Frame = +3
Query: 32 VAEVLGTYFLIFAGCASVVVNLNND---KVVTLPGIAIVWGLAVMVLVYSIGHISGAHFN 88
+AE + T+ ++ +V+ +D K V + GIA +G + LVY ISG H N
Sbjct: 216 IAEFVATFLFLYITILTVMGVNKSDTKCKTVGIQGIAWAFGGMIFALVYCTAGISGGHIN 395
Query: 89 PAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFSGK 131
PAVT ++ L + Y++ QV+G+ +G ++ F GK
Sbjct: 396 PAVTFGLFLARKLSLTRAVFYMVMQVLGAICGAGVVK-GFEGK 521
Score = 33.9 bits (76), Expect = 0.029
Identities = 30/103 (29%), Positives = 48/103 (46%), Gaps = 10/103 (9%)
Frame = +3
Query: 139 LPAGSDLQAFVVEFIITFLLMFV----VSGV---ATDNRAIGELAGL--AVGSTVMLNVL 189
L + S +A + EF+ TFL +++ V GV T + +G + G+ A G + V
Sbjct: 186 LTSWSFYRAGIAEFVATFLFLYITILTVMGVNKSDTKCKTVG-IQGIAWAFGGMIFALVY 362
Query: 190 FAGPITGASMNPARSLGPAIVHN-QYKGIWIYMVSPILGAVAG 231
I+G +NPA + G + YMV +LGA+ G
Sbjct: 363 CTAGISGGHINPAVTFGLFLARKLSLTRAVFYMVMQVLGAICG 491
>TC14565 homologue to UP|O65741 (O65741) Transmembrane channel protein
(Fragment), partial (86%)
Length = 539
Score = 49.3 bits (116), Expect = 7e-07
Identities = 32/81 (39%), Positives = 43/81 (52%), Gaps = 8/81 (9%)
Frame = +2
Query: 156 FLLMFVVSGVATDNRAIGE-----LAGLAVGSTVMLNVLFAGPITGASMNPARSLGPAIV 210
F+L++ V R+ + LA L +G V L L PITG +NPARSLG +IV
Sbjct: 2 FVLVYTVFSATDAKRSARDSHVPILAPLPIGFAVFLVHLATIPITGTGINPARSLGASIV 181
Query: 211 HNQYKG---IWIYMVSPILGA 228
N+ G WI+ V P +GA
Sbjct: 182 FNKDLGWDDHWIFWVGPFIGA 244
>AV409092
Length = 426
Score = 47.8 bits (112), Expect = 2e-06
Identities = 36/110 (32%), Positives = 51/110 (45%), Gaps = 5/110 (4%)
Frame = +3
Query: 25 VPLLQKLVAEVLGTYFLIFAGCASVVV--NLNNDKVVTLPG---IAIVWGLAVMVLVYSI 79
V L+ +AE + T IFAG S + L +D + G +A+ A+ V V
Sbjct: 72 VTSLKAYLAEFIATLLFIFAGVGSAIAYNELTSDAALDPTGLVAVALAHAFALFVGVSIA 251
Query: 80 GHISGAHFNPAVTIAHVTTKRFPLKQVPAYILAQVIGSTLASGTLRLIFS 129
+ISG H NPAVT+ L Y +AQ++GS +A L I S
Sbjct: 252 ANISGGHLNPAVTLGLAVGGNITLLTGFFYWIAQLLGSIVACLLLNFITS 401
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.323 0.138 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,863,309
Number of Sequences: 28460
Number of extensions: 46543
Number of successful extensions: 249
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 227
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 235
length of query: 259
length of database: 4,897,600
effective HSP length: 88
effective length of query: 171
effective length of database: 2,393,120
effective search space: 409223520
effective search space used: 409223520
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0118c.11


