

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
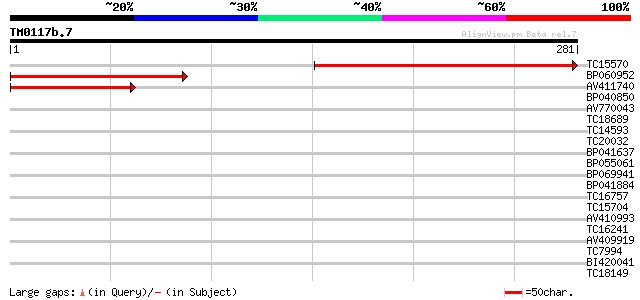
Query= TM0117b.7
(281 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC15570 similar to GB|AAP21363.1|30102890|BT006555 At4g38290 {Ar... 255 7e-69
BP060952 180 3e-46
AV411740 125 6e-30
BP040850 33 0.054
AV770043 27 0.13
TC18689 30 0.46
TC14593 similar to UP|Q8H1U3 (Q8H1U3) CDC26, partial (31%) 28 1.7
TC20032 27 3.0
BP041637 27 3.0
BP055061 27 3.9
BP069941 27 3.9
BP041884 27 5.1
TC16757 weakly similar to UP|Q9SBQ2 (Q9SBQ2) Anthocyanin 5-O-glu... 27 5.1
TC15704 similar to UP|O22265 (O22265) Expressed protein (At2g474... 26 6.6
AV410993 26 6.6
TC16241 weakly similar to UP|Q91TH4 (Q91TH4) T122, partial (7%) 26 6.6
AV409919 26 6.6
TC7994 similar to UP|Q8RU73 (Q8RU73) Ribose-5-phosphate isomeras... 26 8.7
BI420041 26 8.7
TC18149 weakly similar to UP|Q91BL3 (Q91BL3) Essential structura... 26 8.7
>TC15570 similar to GB|AAP21363.1|30102890|BT006555 At4g38290 {Arabidopsis
thaliana;}, partial (41%)
Length = 642
Score = 255 bits (651), Expect = 7e-69
Identities = 130/130 (100%), Positives = 130/130 (100%)
Frame = +1
Query: 152 PSVPISCGKSTMKLAPVESNCVTTSSGPTIGNPKGLIDMQWEERYQQLQMLLRKLDQSDQ 211
PSVPISCGKSTMKLAPVESNCVTTSSGPTIGNPKGLIDMQWEERYQQLQMLLRKLDQSDQ
Sbjct: 1 PSVPISCGKSTMKLAPVESNCVTTSSGPTIGNPKGLIDMQWEERYQQLQMLLRKLDQSDQ 180
Query: 212 VEYVQMLQSLSSVELSRHAVELEKRAIQLSLEEAKELQRVAVLNVLGKSAKKFKAPADHD 271
VEYVQMLQSLSSVELSRHAVELEKRAIQLSLEEAKELQRVAVLNVLGKSAKKFKAPADHD
Sbjct: 181 VEYVQMLQSLSSVELSRHAVELEKRAIQLSLEEAKELQRVAVLNVLGKSAKKFKAPADHD 360
Query: 272 ECSDKLKTSS 281
ECSDKLKTSS
Sbjct: 361 ECSDKLKTSS 390
>BP060952
Length = 545
Score = 180 bits (456), Expect = 3e-46
Identities = 88/88 (100%), Positives = 88/88 (100%)
Frame = +3
Query: 1 MDLPNRDKQPTVDVKKTASRDLQNDNKIMALTSVGSSSVLKDKDPDTDSNRISGTKRPLS 60
MDLPNRDKQPTVDVKKTASRDLQNDNKIMALTSVGSSSVLKDKDPDTDSNRISGTKRPLS
Sbjct: 282 MDLPNRDKQPTVDVKKTASRDLQNDNKIMALTSVGSSSVLKDKDPDTDSNRISGTKRPLS 461
Query: 61 DYPVNHHLQQSPGNSTANGHLVYVRRKS 88
DYPVNHHLQQSPGNSTANGHLVYVRRKS
Sbjct: 462 DYPVNHHLQQSPGNSTANGHLVYVRRKS 545
>AV411740
Length = 412
Score = 125 bits (315), Expect = 6e-30
Identities = 62/62 (100%), Positives = 62/62 (100%)
Frame = +1
Query: 1 MDLPNRDKQPTVDVKKTASRDLQNDNKIMALTSVGSSSVLKDKDPDTDSNRISGTKRPLS 60
MDLPNRDKQPTVDVKKTASRDLQNDNKIMALTSVGSSSVLKDKDPDTDSNRISGTKRPLS
Sbjct: 226 MDLPNRDKQPTVDVKKTASRDLQNDNKIMALTSVGSSSVLKDKDPDTDSNRISGTKRPLS 405
Query: 61 DY 62
DY
Sbjct: 406 DY 411
>BP040850
Length = 588
Score = 33.1 bits (74), Expect = 0.054
Identities = 41/149 (27%), Positives = 62/149 (41%), Gaps = 9/149 (6%)
Frame = +1
Query: 60 SDYPVNHHLQQSPGNSTA---NGHLVYVRRKSEAELGKCAAFENP--TNNAFCQPPRQLC 114
S Y +HH Q ST+ + HL S C E P T +A C P R L
Sbjct: 124 SRYL*SHHSLQLHTPSTSLRRDNHLSPTHLTSSPSEPPCCLPEKPPPTTSAACSPGRVLM 303
Query: 115 SEEETALPKPQLKEPNVSCFPAFAPFPLTSSISSSGRPSVPISCGKSTMKL--APVESNC 172
S + P P L P S F + PF +++SS +P P++ +++ +P ++
Sbjct: 304 SFVTPSYPSPSL-APMPSSFSSLNPF---KAVTSSPKP-YPLARSRTSRIC*NSPWKAGR 468
Query: 173 VTTS--SGPTIGNPKGLIDMQWEERYQQL 199
+T S S P KG +W+ L
Sbjct: 469 LTPSPLSSPPAETRKGSR*FRWQSELGSL 555
>AV770043
Length = 532
Score = 27.3 bits (59), Expect(2) = 0.13
Identities = 15/51 (29%), Positives = 25/51 (48%)
Frame = -1
Query: 103 NNAFCQPPRQLCSEEETALPKPQLKEPNVSCFPAFAPFPLTSSISSSGRPS 153
N++ P ++LC + +P S +P+ P P +SS SSG P+
Sbjct: 154 NSSIHPPNKRLCLQFSHRIPSSTR*PHPQSSYPSHPPAPESSSPCSSGGPT 2
Score = 23.1 bits (48), Expect(2) = 0.13
Identities = 8/16 (50%), Positives = 10/16 (62%)
Frame = -3
Query: 57 RPLSDYPVNHHLQQSP 72
+P YP+ HHL Q P
Sbjct: 296 KPPPSYPLPHHLPQMP 249
>TC18689
Length = 397
Score = 30.0 bits (66), Expect = 0.46
Identities = 19/50 (38%), Positives = 25/50 (50%)
Frame = -2
Query: 129 PNVSCFPAFAPFPLTSSISSSGRPSVPISCGKSTMKLAPVESNCVTTSSG 178
PN P F P SSI S+ +PI C +T + + SN TTS+G
Sbjct: 189 PNPHARPMFQP---PSSIMSTSCELLPIQCCTTTTTIRGLYSNYKTTSNG 49
>TC14593 similar to UP|Q8H1U3 (Q8H1U3) CDC26, partial (31%)
Length = 1112
Score = 28.1 bits (61), Expect = 1.7
Identities = 24/70 (34%), Positives = 29/70 (41%), Gaps = 7/70 (10%)
Frame = +3
Query: 100 NPTNNA-FCQPPRQLCSEEETALPKPQLKEPNVSCFPAFAPFPLTSSISSSGRP------ 152
NP+ +A F P S +A P P+L + C P P S ISSS P
Sbjct: 210 NPSPSAPFLSPLPIWRSRSSSASPTPKLTAKSPPCSPPSTSPPTASIISSSTAPPPSSPQ 389
Query: 153 SVPISCGKST 162
S P S ST
Sbjct: 390 SAPFSGSDST 419
>TC20032
Length = 655
Score = 27.3 bits (59), Expect = 3.0
Identities = 15/37 (40%), Positives = 21/37 (56%)
Frame = -3
Query: 143 TSSISSSGRPSVPISCGKSTMKLAPVESNCVTTSSGP 179
T+ +S SG S P +C KST +A S C + S+ P
Sbjct: 146 TAGVSGSGSES-PAACRKSTASVAWKLSTCPSPSASP 39
>BP041637
Length = 457
Score = 27.3 bits (59), Expect = 3.0
Identities = 15/30 (50%), Positives = 19/30 (63%), Gaps = 2/30 (6%)
Frame = +1
Query: 134 FPAFAPFPLTSSISSSGRPSVP--ISCGKS 161
F +F+ F T S+SSS PS+P IS KS
Sbjct: 364 FTSFSSFTFTFSLSSSPSPSIPSLISLSKS 453
>BP055061
Length = 544
Score = 26.9 bits (58), Expect = 3.9
Identities = 14/31 (45%), Positives = 18/31 (57%)
Frame = -3
Query: 192 WEERYQQLQMLLRKLDQSDQVEYVQMLQSLS 222
W+E + LRKL S QV +QMLQ +S
Sbjct: 389 WKEWRKS*HQKLRKLKLSPQVVLIQMLQKVS 297
>BP069941
Length = 304
Score = 26.9 bits (58), Expect = 3.9
Identities = 9/22 (40%), Positives = 14/22 (62%)
Frame = -2
Query: 101 PTNNAFCQPPRQLCSEEETALP 122
P N +FC PR++C E+ +P
Sbjct: 297 PLNRSFCMIPRKVCCEKARFIP 232
>BP041884
Length = 489
Score = 26.6 bits (57), Expect = 5.1
Identities = 22/67 (32%), Positives = 29/67 (42%), Gaps = 7/67 (10%)
Frame = +3
Query: 112 QLCSEEETALPKPQLKE--PNVSCFP---AFAPFPLTSSISSSGRPS--VPISCGKSTMK 164
QLC P P +K + +C P A AP P+T IS+ RPS P+
Sbjct: 6 QLCKNPR---PNPDIKTLFTDHTCSPPNGAHAPTPVTLPISAGARPSFFAPLGAHGGPFT 176
Query: 165 LAPVESN 171
AP +N
Sbjct: 177 PAPPAAN 197
>TC16757 weakly similar to UP|Q9SBQ2 (Q9SBQ2) Anthocyanin
5-O-glucosyltransferase, partial (19%)
Length = 638
Score = 26.6 bits (57), Expect = 5.1
Identities = 15/35 (42%), Positives = 19/35 (53%), Gaps = 4/35 (11%)
Frame = -3
Query: 115 SEEETALPKPQLKEPNVSCF----PAFAPFPLTSS 145
SEE + P K N S F AF+PFP+T+S
Sbjct: 273 SEEPPSFPASIAKPLNFSAFLLTSSAFSPFPITAS 169
>TC15704 similar to UP|O22265 (O22265) Expressed protein
(At2g47450/T30B22.25), partial (25%)
Length = 664
Score = 26.2 bits (56), Expect = 6.6
Identities = 18/48 (37%), Positives = 24/48 (49%)
Frame = -3
Query: 134 FPAFAPFPLTSSISSSGRPSVPISCGKSTMKLAPVESNCVTTSSGPTI 181
+ F+PFPL S+ISS+ S AP S +TT S PT+
Sbjct: 152 YSKFSPFPLRSNISSTSAYS----------NTAP--SKTLTTPSSPTL 45
>AV410993
Length = 427
Score = 26.2 bits (56), Expect = 6.6
Identities = 28/92 (30%), Positives = 36/92 (38%), Gaps = 1/92 (1%)
Frame = -2
Query: 68 LQQSPGNSTANGHLVYVRRKSEAELGKCAAFENPTNNAFCQPPRQLCSEEETALPKPQLK 127
L ++P S N ++ + + + C F NP N AF P K
Sbjct: 249 LSENPRRSMRNFFILNIFPFTSSISFICGCF-NPINRAFFSP-----------------K 124
Query: 128 EPNV-SCFPAFAPFPLTSSISSSGRPSVPISC 158
P+ S F FA F T SISS R ISC
Sbjct: 123 FPSTASIFFFFAKFSSTGSISSRDRIIDAISC 28
>TC16241 weakly similar to UP|Q91TH4 (Q91TH4) T122, partial (7%)
Length = 799
Score = 26.2 bits (56), Expect = 6.6
Identities = 21/83 (25%), Positives = 31/83 (37%)
Frame = +1
Query: 102 TNNAFCQPPRQLCSEEETALPKPQLKEPNVSCFPAFAPFPLTSSISSSGRPSVPISCGKS 161
+N+ QPP T + P + S +P+ P L SIS+S P S
Sbjct: 184 SNSPTSQPPVAQTPRAVTPISSPSSSPTSNSPYPSTNPPSLAPSISTSPPALTPAS---- 351
Query: 162 TMKLAPVESNCVTTSSGPTIGNP 184
PV + S P+ +P
Sbjct: 352 ----TPVPATASPPSQSPSAESP 408
>AV409919
Length = 423
Score = 26.2 bits (56), Expect = 6.6
Identities = 13/47 (27%), Positives = 20/47 (41%)
Frame = +3
Query: 109 PPRQLCSEEETALPKPQLKEPNVSCFPAFAPFPLTSSISSSGRPSVP 155
PP + LP P L P + P+ +P +SS + + R P
Sbjct: 108 PPANASKPRTSKLPPPPLPSPTLYFSPSSSPSSTSSSTAGARRFGTP 248
>TC7994 similar to UP|Q8RU73 (Q8RU73) Ribose-5-phosphate isomerase
precursor , partial (81%)
Length = 1355
Score = 25.8 bits (55), Expect = 8.7
Identities = 23/87 (26%), Positives = 32/87 (36%)
Frame = +2
Query: 90 AELGKCAAFENPTNNAFCQPPRQLCSEEETALPKPQLKEPNVSCFPAFAPFPLTSSISSS 149
AEL C P+++ PP T +P P L P PL +
Sbjct: 236 AELEFCQWLPYPSHHHHLSPPH-------TTMPPPTLSYA--------PPLPLNYAPPPL 370
Query: 150 GRPSVPISCGKSTMKLAPVESNCVTTS 176
PSVP K+T + +P T+S
Sbjct: 371 PNPSVPSPSPKTTSRDSPPTRPSTTSS 451
>BI420041
Length = 452
Score = 25.8 bits (55), Expect = 8.7
Identities = 16/58 (27%), Positives = 24/58 (40%)
Frame = -3
Query: 135 PAFAPFPLTSSISSSGRPSVPISCGKSTMKLAPVESNCVTTSSGPTIGNPKGLIDMQW 192
P P P ++SSS + SC S+ +P S+ + SS + ID W
Sbjct: 405 PPEPPSPA*GTLSSSSSSNSSSSCSSSS*YSSPSSSSSFSGSS*SNSSSSSSSIDTSW 232
>TC18149 weakly similar to UP|Q91BL3 (Q91BL3) Essential structural protein
pp78/81, partial (5%)
Length = 710
Score = 25.8 bits (55), Expect = 8.7
Identities = 12/33 (36%), Positives = 18/33 (54%)
Frame = +2
Query: 123 KPQLKEPNVSCFPAFAPFPLTSSISSSGRPSVP 155
K L +PN SC + P T SIS + + ++P
Sbjct: 182 KSILSDPNPSCSSERSILPTTCSISGNPKSAIP 280
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.310 0.127 0.356
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,714,866
Number of Sequences: 28460
Number of extensions: 71634
Number of successful extensions: 346
Number of sequences better than 10.0: 41
Number of HSP's better than 10.0 without gapping: 344
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 346
length of query: 281
length of database: 4,897,600
effective HSP length: 89
effective length of query: 192
effective length of database: 2,364,660
effective search space: 454014720
effective search space used: 454014720
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0117b.7


