

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0117b.11
(669 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
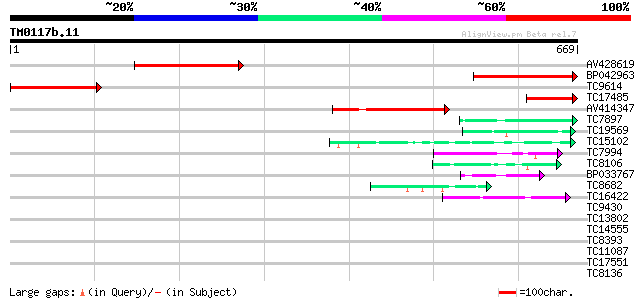
Score E
Sequences producing significant alignments: (bits) Value
AV428619 244 4e-65
BP042963 235 2e-62
TC9614 similar to UP|O22799 (O22799) At2g33490 protein, partial ... 218 2e-57
TC17485 114 4e-26
AV414347 100 6e-22
TC7897 weakly similar to UP|ENL2_ARATH (Q9T076) Early nodulin-li... 45 4e-05
TC19569 similar to UP|AAR11301 (AAR11301) Lectin-like receptor k... 45 4e-05
TC15102 similar to UP|Q7X9B3 (Q7X9B3) 9/13 hydroperoxide lyase, ... 44 6e-05
TC7994 similar to UP|Q8RU73 (Q8RU73) Ribose-5-phosphate isomeras... 43 1e-04
TC8106 weakly similar to UP|Q9XIV1 (Q9XIV1) mRNA expressed in cu... 42 2e-04
BP033767 42 3e-04
TC8682 similar to UP|HPPD_SOLSC (Q9ARF9) 4-hydroxyphenylpyruvate... 42 3e-04
TC16422 similar to PIR|T05653|T05653 amino acid transport protei... 41 7e-04
TC9430 similar to UP|Q96569 (Q96569) L-lactate dehydrogenase (L... 40 0.001
TC13802 similar to UP|Q9M1T1 (Q9M1T1) Sugar-phosphate isomerase-... 40 0.001
TC14555 similar to UP|Q43558 (Q43558) Proline rich protein precu... 40 0.001
TC8393 weakly similar to GB|AAM61124.1|21536792|AY084557 prenyla... 40 0.002
TC11087 weakly similar to UP|Q8GS92 (Q8GS92) OJ1513_F02.5 protei... 39 0.002
TC17551 weakly similar to UP|AAR24662 (AAR24662) At5g26960, part... 39 0.002
TC8136 similar to GB|AAB60729.1|2160166|F21M12 F21M12.13 {Arabid... 38 0.005
>AV428619
Length = 405
Score = 244 bits (622), Expect = 4e-65
Identities = 125/128 (97%), Positives = 125/128 (97%)
Frame = +1
Query: 148 RSHIIQTITIPSESLLNELRIVEEMKRQCDEKRDVYEYMATRFRERGRSKGGKTETFSLQ 207
RSHIIQTITIPSESLLNELRIVEEMKRQCDEKRDVYEYMATRFRERGRSKGGK ETFSLQ
Sbjct: 22 RSHIIQTITIPSESLLNELRIVEEMKRQCDEKRDVYEYMATRFRERGRSKGGKRETFSLQ 201
Query: 208 QLQTARDEYDEEATLFVFRLKSLKQGQSRSLLTQAARHHAAQLCFFKKAVKSLETVEPHV 267
QLQTARDEYDEEATLFVFRLKSLKQGQSRSLLTQAARHHAAQL FKKAVKSLETVEPHV
Sbjct: 202 QLQTARDEYDEEATLFVFRLKSLKQGQSRSLLTQAARHHAAQLWVFKKAVKSLETVEPHV 381
Query: 268 KSVTEQQH 275
KSVTEQQH
Sbjct: 382 KSVTEQQH 405
>BP042963
Length = 517
Score = 235 bits (599), Expect = 2e-62
Identities = 121/122 (99%), Positives = 121/122 (99%)
Frame = -1
Query: 548 ASPTIVSSPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAAS 607
ASPTIVSSPKISELHELPRPPTNFPS SRLLGLVGYSGPLVPRGQKVSAPNNLVISSAAS
Sbjct: 517 ASPTIVSSPKISELHELPRPPTNFPSXSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAAS 338
Query: 608 PLPIPPQAMARSFSIPSSGARVTALHSSSALESSHISSISEDIASPPLTPIALSNSRPSS 667
PLPIPPQAMARSFSIPSSGARVTALHSSSALESSHISSISEDIASPPLTPIALSNSRPSS
Sbjct: 337 PLPIPPQAMARSFSIPSSGARVTALHSSSALESSHISSISEDIASPPLTPIALSNSRPSS 158
Query: 668 DG 669
DG
Sbjct: 157 DG 152
>TC9614 similar to UP|O22799 (O22799) At2g33490 protein, partial (12%)
Length = 673
Score = 218 bits (555), Expect = 2e-57
Identities = 108/108 (100%), Positives = 108/108 (100%)
Frame = +2
Query: 1 MKSSLKKLRGLGLHNHNHNHKHHDSKTILPLGQLEELAQATRDMQDMRDCYDTLLSAAAA 60
MKSSLKKLRGLGLHNHNHNHKHHDSKTILPLGQLEELAQATRDMQDMRDCYDTLLSAAAA
Sbjct: 233 MKSSLKKLRGLGLHNHNHNHKHHDSKTILPLGQLEELAQATRDMQDMRDCYDTLLSAAAA 412
Query: 61 TASSAYEFAESLRDMGSCLLEKTALNDHEEETGKVLLMLGKIQFKLQK 108
TASSAYEFAESLRDMGSCLLEKTALNDHEEETGKVLLMLGKIQFKLQK
Sbjct: 413 TASSAYEFAESLRDMGSCLLEKTALNDHEEETGKVLLMLGKIQFKLQK 556
Score = 35.8 bits (81), Expect = 0.023
Identities = 22/40 (55%), Positives = 23/40 (57%)
Frame = +3
Query: 129 FCFNFITNFSVFLFFQFAQRSHIIQTITIPSESLLNELRI 168
FC + S F RSHIIQTITIPS SLL EL I
Sbjct: 516 FCSCLAKSSSNFRKLIDKYRSHIIQTITIPSXSLLXELPI 635
>TC17485
Length = 462
Score = 114 bits (286), Expect = 4e-26
Identities = 59/59 (100%), Positives = 59/59 (100%)
Frame = +2
Query: 611 IPPQAMARSFSIPSSGARVTALHSSSALESSHISSISEDIASPPLTPIALSNSRPSSDG 669
IPPQAMARSFSIPSSGARVTALHSSSALESSHISSISEDIASPPLTPIALSNSRPSSDG
Sbjct: 2 IPPQAMARSFSIPSSGARVTALHSSSALESSHISSISEDIASPPLTPIALSNSRPSSDG 178
>AV414347
Length = 424
Score = 100 bits (250), Expect = 6e-22
Identities = 58/141 (41%), Positives = 88/141 (62%), Gaps = 3/141 (2%)
Frame = +2
Query: 381 SEKLRQMRPSLSRKFSSYVLPTPVDAKSSISSGSNNPKPSKMQTNSNEATTNLWHSSPLE 440
+EK+R + S + K ++YVLPTPV+ K + +S S + +++ ++ NLWHSSPL+
Sbjct: 5 AEKVRLLLSSSAAKPNAYVLPTPVNIKETKTS-------SAPRLSASGSSHNLWHSSPLD 163
Query: 441 QKKHET--IRDEFSSPTVRNAQSVLKESNSNTATTRLPPPLVDGLLSSNHDYVSAY-SKK 497
+KK+E + + S P + A S+LKESNS+ + +LP P +GL D +A SKK
Sbjct: 164 EKKNEKDLVDGKLSEPAIPRAHSILKESNSDNTSIQLPRPSAEGLSLPQFDIFNASDSKK 343
Query: 498 IKRHAFSGPLTSNPMPTRPVS 518
R AFSGPLT+ P+ +PVS
Sbjct: 344 TLRQAFSGPLTNKPLSVKPVS 406
>TC7897 weakly similar to UP|ENL2_ARATH (Q9T076) Early nodulin-like protein
2 precursor (Phytocyanin-like protein), partial (31%)
Length = 1482
Score = 45.1 bits (105), Expect = 4e-05
Identities = 36/139 (25%), Positives = 51/139 (35%)
Frame = +3
Query: 531 PTPIPQPPSSSPKVSPSASPTIVSSPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPR 590
P P P PP SSP SP ASP S P E PP+ PS + P+
Sbjct: 528 PPPSP-PPKSSPSPSPKASPPSGSPPTAGEPSPAGTPPSVLPSPA--------GAPVPAG 680
Query: 591 GQKVSAPNNLVISSAASPLPIPPQAMARSFSIPSSGARVTALHSSSALESSHISSISEDI 650
G P + S + SP+ PP A + S + + + +S +
Sbjct: 681 GPSAGTPPSASTSPSPSPVSAPPAASPAPGAESPSSSPTSNSPAGGPTATSPSPAGDSPA 860
Query: 651 ASPPLTPIALSNSRPSSDG 669
PP ++ PS+ G
Sbjct: 861 GGPPAPSPTTGDTPPSASG 917
Score = 33.5 bits (75), Expect = 0.11
Identities = 34/137 (24%), Positives = 57/137 (40%), Gaps = 6/137 (4%)
Frame = +3
Query: 511 PMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSP---KVSPSASPTIVSSPKISELHELPRP 567
P+P S + S P+P+ PP++SP SPS+SPT +SP P P
Sbjct: 666 PVPAGGPSAGTPPSASTSPSPSPVSAPPAASPAPGAESPSSSPT-SNSPAGGPTATSPSP 842
Query: 568 PTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLPIP---PQAMARSFSIPS 624
+ P+ G P G + + +S + +P +P P + + + + P
Sbjct: 843 AGDSPAG-------GPPAPSPTTGDTPPSASGPDVSPSGTPSSLPADTPSSSSNNSTAPP 1001
Query: 625 SGARVTALHSSSALESS 641
G +++ SS L S
Sbjct: 1002PGNGGSSVVPSSFLHYS 1052
>TC19569 similar to UP|AAR11301 (AAR11301) Lectin-like receptor kinase 1;1,
partial (16%)
Length = 486
Score = 45.1 bits (105), Expect = 4e-05
Identities = 41/136 (30%), Positives = 53/136 (38%), Gaps = 3/136 (2%)
Frame = +1
Query: 535 PQPPSSSPKVSPSASPTIVSSPKISELHELPRPPTNFPSNSRLLGLVGYS---GPLVPRG 591
P PPS P SPS SPT ++P S PPT PS S S P P
Sbjct: 76 PPPPSQQPTHSPSTSPTSTTTPPPSPTKVTENPPT-APSTSTKSPTSSASEEPSPPNPST 252
Query: 592 QKVSAPNNLVISSAASPLPIPPQAMARSFSIPSSGARVTALHSSSALESSHISSISEDIA 651
P + S ASP P PP + S + ++A S ++ +S +
Sbjct: 253 STTQQPKHSPTSPPASPSPSPPSTKPPTAMASPSTSPLSATKSHPTPPAAPSASST---- 420
Query: 652 SPPLTPIALSNSRPSS 667
PP T + S PSS
Sbjct: 421 PPPTTTSLKTMSSPSS 468
Score = 37.7 bits (86), Expect = 0.006
Identities = 42/151 (27%), Positives = 61/151 (39%), Gaps = 7/151 (4%)
Frame = +1
Query: 502 AFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKVSPSASPTIVSSPKISEL 561
++S P + PT S + P PT + + P ++P S + SPT S
Sbjct: 64 SYSPPPPPSQQPTHSPSTSPTSTTTPPPSPTKVTENPPTAPSTS-TKSPT-------SSA 219
Query: 562 HELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSA-PNNLVISSAASPL------PIPPQ 614
E P PP S ++ S P P S P + S + SPL P PP
Sbjct: 220 SEEPSPPNPSTSTTQQPKHSPTSPPASPSPSPPSTKPPTAMASPSTSPLSATKSHPTPPA 399
Query: 615 AMARSFSIPSSGARVTALHSSSALESSHISS 645
A + S + P + T+L + S+ SS SS
Sbjct: 400 APSASSTPPPT---TTSLKTMSSPSSSTPSS 483
>TC15102 similar to UP|Q7X9B3 (Q7X9B3) 9/13 hydroperoxide lyase, partial
(68%)
Length = 1097
Score = 44.3 bits (103), Expect = 6e-05
Identities = 75/305 (24%), Positives = 114/305 (36%), Gaps = 15/305 (4%)
Frame = +1
Query: 378 PDSSEKLRQ------MRPSLSRKFSSYVLPTPVDAKSSI---------SSGSNNPKPSKM 422
P EK +Q +R S +++ ++ TP A ++ +S S+ P +
Sbjct: 7 PSQFEKTKQTNRYSSVRSSHQQQWQQHLQTTPKTAAATFL*NPSLEATASPSSEPCTTAT 186
Query: 423 QTNSNEATTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTATTRLPPPLVDG 482
T++ +A TN SSP E + E + + K S+S+T PPP
Sbjct: 187 TTSTTKAATN---SSPPESRSTTPPSSEPTCLLAPASPPTPKSSHSST-----PPP---- 330
Query: 483 LLSSNHDYVSAYSKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSP 542
S + SK +A S S P P P + + P P PP++S
Sbjct: 331 ---SRSSSTTPKSK----NATSLTAPSCPPPASPAA-------TAPAPSKTPPNPPTNSS 468
Query: 543 KVSPSASPTIVSSPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVI 602
K PS+ + + + S L P PT P S P P Q+
Sbjct: 469 K--PSSCKSSLPNTTPSSLSSAPPSPTTSP----------ISTPSSPENQE--------- 585
Query: 603 SSAASPLPIPPQAMARSFSIPSSGARVTALHSSSALESSHISSISEDIASPPLTPIALSN 662
P PP A RS S SS T SSA ++ +SPP + ++L +
Sbjct: 586 ----KPASTPPSAPPRSTSSSSSSPINTPRKPSSATKA--------PASSPPGSELSLHH 729
Query: 663 SRPSS 667
S PS+
Sbjct: 730 SPPSA 744
>TC7994 similar to UP|Q8RU73 (Q8RU73) Ribose-5-phosphate isomerase
precursor , partial (81%)
Length = 1355
Score = 43.1 bits (100), Expect = 1e-04
Identities = 46/161 (28%), Positives = 67/161 (41%), Gaps = 9/161 (5%)
Frame = +2
Query: 501 HAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTP-IPQP-PSSSPKVSPSASPTIVSSPKI 558
H S P T+ P PT + ++ P LP P +P P P ++ + SP P+ SSP
Sbjct: 281 HHLSPPHTTMPPPTLSYAPPLPLNYAPPPLPNPSVPSPSPKTTSRDSPPTRPSTTSSPAW 460
Query: 559 SELHE-LPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLPIPPQAMA 617
S P PP++ PS++ S P N SSA+ PP +
Sbjct: 461 SSASAPAPPPPSSSPSSA-----------------SFSPPANSPTSSAS-----PPPSAR 574
Query: 618 RS------FSIPSSGARVTALHSSSALESSHISSISEDIAS 652
RS PSS ++ S+A S +SS S +A+
Sbjct: 575 RSRRDPSASRSPSSTTIPASISPSTAPTRSTLSSTSSRVAA 697
>TC8106 weakly similar to UP|Q9XIV1 (Q9XIV1) mRNA expressed in cucumber
hypocotyls (Arabinogalactan protein), partial (32%)
Length = 760
Score = 42.4 bits (98), Expect = 2e-04
Identities = 42/156 (26%), Positives = 58/156 (36%), Gaps = 3/156 (1%)
Frame = +1
Query: 499 KRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKVSPSASPTIVSSPKI 558
K A P+ P T P +V + S P P P+ PP+ P SP A +P +
Sbjct: 55 KAAAAPAPVAKAPASTPPAAVP---VSSPPPAPVPVASPPAPVPVSSPPAPAPTTPAPAV 225
Query: 559 SELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPL---PIPPQA 615
+ E+P P PS S+ V +G+K AP A SPL P PP
Sbjct: 226 TPTAEVPAPA---PSKSK---------KKVKKGKKHGAP-------APSPLLGPPAPPVG 348
Query: 616 MARSFSIPSSGARVTALHSSSALESSHISSISEDIA 651
S G A S A ++ + +A
Sbjct: 349 APGPDDALSPGPTAVAADESGAETIRYLKMVVGSLA 456
>BP033767
Length = 541
Score = 42.0 bits (97), Expect = 3e-04
Identities = 36/100 (36%), Positives = 40/100 (40%), Gaps = 1/100 (1%)
Frame = +1
Query: 533 PIPQPPSSSPKVSPSASPTIVSSPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQ 592
P PQPP P P +SSP H L PP + PS P P
Sbjct: 250 PPPQPP-------PPKMPPHLSSPHTHLHHHLLLPPPSPPSP-----------PTSPPSS 375
Query: 593 KVSAPN-NLVISSAASPLPIPPQAMARSFSIPSSGARVTA 631
+ PN N SS+ASPLP P S S PSS A A
Sbjct: 376 SLKPPNPNPPTSSSASPLPPSPAPSPSSLSPPSSTADAAA 495
Score = 30.4 bits (67), Expect = 0.96
Identities = 26/93 (27%), Positives = 39/93 (40%), Gaps = 10/93 (10%)
Frame = +3
Query: 504 SGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKVSPSASPTI-----VSSPKI 558
S P+ + + +P + S+ P P P P P +PK S++PT S+P
Sbjct: 117 SEPIINRRILHQPFTQGSLPPSQPPNPPPPSPSPSPPNPKYPFSSTPTTTTTQNASTPFF 296
Query: 559 SELHELPRPPT-----NFPSNSRLLGLVGYSGP 586
P PP+ +FP+N L L S P
Sbjct: 297 PTYPSPPPPPSPSSFASFPANISSLILPQTSKP 395
Score = 30.0 bits (66), Expect = 1.3
Identities = 30/141 (21%), Positives = 51/141 (35%), Gaps = 8/141 (5%)
Frame = +1
Query: 424 TNSNEATTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTATTRLPPPLVDGL 483
+++ ++TN H +P +T P ++ ++ + PPP +
Sbjct: 127 SSTGGSSTNPSHKAPFHPLNPQTHHHHHHLPHLQT------QNTHFPPPPQPPPPKMPPH 288
Query: 484 LSSNHDYVSAYSKKIKRHAFSGPLTSNPMPTRPVSVD--------SVQMFSGPLLPTPIP 535
LSS H + + H P + PT P S + PL P+P P
Sbjct: 289 LSSPHTH-------LHHHLLLPPPSPPSPPTSPPSSSLKPPNPNPPTSSSASPLPPSPAP 447
Query: 536 QPPSSSPKVSPSASPTIVSSP 556
P S SP S + + + P
Sbjct: 448 SPSSLSPPSSTADAAAKTTPP 510
>TC8682 similar to UP|HPPD_SOLSC (Q9ARF9) 4-hydroxyphenylpyruvate
dioxygenase (4HPPD) (HPD) (HPPDase) , partial (29%)
Length = 576
Score = 42.0 bits (97), Expect = 3e-04
Identities = 48/178 (26%), Positives = 68/178 (37%), Gaps = 35/178 (19%)
Frame = +1
Query: 426 SNEATTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSVLKESN------------------ 467
S+ AT + +SP H TIR+ +P+ N Q +LK+S
Sbjct: 52 SHPATCHSAATSPT*HHHHPTIRNPTLTPSWVNKQQLLKKSKPGSNSSGSTASSGPTQNQ 231
Query: 468 -------SNTATTRLPPPLVDGLLS-------SNHDYVSAYSKKIKRHAFSGPLTS---N 510
S T+++ P P S S+ + S + S P TS +
Sbjct: 232 TGSP*KGSTTSSSGAPTPPTPHAASPGASACRSSQNQTSPPETTLTPPTSSAPATSASSS 411
Query: 511 PMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKVSPSASPTIVSSPKISELHELPRPP 568
P T P S S P P+P+ PP +SP P+ASP+ SP S P PP
Sbjct: 412 PPHTLPKSP------SPPPPPSPLSPPPPASPSPPPTASPS-APSPSKSPTPSKPSPP 564
>TC16422 similar to PIR|T05653|T05653 amino acid transport protein homolog
F22I13.20 - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (38%)
Length = 685
Score = 40.8 bits (94), Expect = 7e-04
Identities = 46/155 (29%), Positives = 65/155 (41%), Gaps = 4/155 (2%)
Frame = +3
Query: 511 PMPTRPVSVDSVQMFSGPLL--PTPIPQPPSSSPKVSPSASPTIVSSPKISELHELPRPP 568
P P+ P + P L P P P SSSP +P +S ++ +S SE P
Sbjct: 120 PTPSHPTLEKTHLSSPNPHLSPPNSKPSPTSSSPSSAPESSASLTAS---SEPAGSPASS 290
Query: 569 TNFPSNSRLLGLVGYSGPLVPRG-QKVSAPNNLVISSAASPLPIPPQAMARSFSIPSSG- 626
+ PS S S PL + PN+ +++ASP + ARS + PS+
Sbjct: 291 CSSPSPSSPTTA*CSSSPLAASSIPSLGTPNSDPSATSASP------SAARSAASPSTP* 452
Query: 627 ARVTALHSSSALESSHISSISEDIASPPLTPIALS 661
L S+SA SS + A+PP TP S
Sbjct: 453 LSSPRLDSASATSSSSRPLSASSPATPPATPCRFS 557
>TC9430 similar to UP|Q96569 (Q96569) L-lactate dehydrogenase (LDH) ,
partial (49%)
Length = 866
Score = 40.4 bits (93), Expect = 0.001
Identities = 44/155 (28%), Positives = 69/155 (44%), Gaps = 11/155 (7%)
Frame = +1
Query: 526 SGPLLPTPIPQPPSSSPKVSPSASPTI-------VSSPKISELHELPRPPTNFPSNSRLL 578
S P P+P PPS++P+ SPS++P SSPK S + LP TN +
Sbjct: 292 SNPSPTPPLPPPPSATPR-SPSSAPATSAWPSLKPSSPKNSTKNALPSSTTNRTTPGGKG 468
Query: 579 GLVGYSGPLVPRGQKVSAPNNLVISSAASPLPI---PPQAMARSFSIP-SSGARVTALHS 634
P +PR + L+ ++ + P PI P +ARS + P S+ R T+ S
Sbjct: 469 STSRRPPPFLPR----TRIQRLLSTTLSLPAPISASSPPELARSPASPGSTSCRGTSPSS 636
Query: 635 SSALESSHISSISEDIASPPLTPIALSNSRPSSDG 669
S + ++ + +S P ++ S SS G
Sbjct: 637 SRSYPLWFVTPRTVSSSSFPTPSMSSPTSLGSSPG 741
>TC13802 similar to UP|Q9M1T1 (Q9M1T1) Sugar-phosphate isomerase-like
protein, partial (41%)
Length = 522
Score = 40.4 bits (93), Expect = 0.001
Identities = 35/105 (33%), Positives = 47/105 (44%), Gaps = 8/105 (7%)
Frame = +2
Query: 531 PTPIPQPPSSSPKVSPSASP------TIVSSPKI--SELHELPRPPTNFPSNSRLLGLVG 582
PT P+P S S PS++P T SSP S P PPT+F + SR L L
Sbjct: 236 PTKSPKPSSPSASAPPSSTPLTPSTATSESSPTATSSSSSASPAPPTSFSALSRALELKA 415
Query: 583 YSGPLVPRGQKVSAPNNLVISSAASPLPIPPQAMARSFSIPSSGA 627
+ S+P S +SP P PP A+ S S+ S+ +
Sbjct: 416 LA----------SSP-----SPPSSPPPSPPSAICMSTSLSSASS 505
Score = 30.8 bits (68), Expect = 0.74
Identities = 34/143 (23%), Positives = 54/143 (36%)
Frame = +2
Query: 525 FSGPLLPTPIPQPPSSSPKVSPSASPTIVSSPKISELHELPRPPTNFPSNSRLLGLVGYS 584
F +LP + P S++P +S S P+P + +S L
Sbjct: 32 FQSTILPNKPLHSTLTKPPSQISSNPNKTTSTTSSPTSTTPKPSPSPAPSSTLRAPSSSP 211
Query: 585 GPLVPRGQKVSAPNNLVISSAASPLPIPPQAMARSFSIPSSGARVTALHSSSALESSHIS 644
P +P S++A PP + + S +S + TA SSS+ + +
Sbjct: 212 ASASPASSPTKSPKPSSPSASA-----PPSSTPLTPSTATSESSPTATSSSSSASPAPPT 376
Query: 645 SISEDIASPPLTPIALSNSRPSS 667
S S + L +A S S PSS
Sbjct: 377 SFSALSRALELKALASSPSPPSS 445
>TC14555 similar to UP|Q43558 (Q43558) Proline rich protein precursor,
partial (43%)
Length = 1038
Score = 40.4 bits (93), Expect = 0.001
Identities = 49/176 (27%), Positives = 66/176 (36%)
Frame = +2
Query: 461 SVLKESNSNTATTRLPPPLVDGLLSSNHDYVSAYSKKIKRHAFSGPLTSNPMPTRPVSVD 520
SV +S SN+ TT PP + +A S + P+ S+P P S
Sbjct: 173 SVGAQSPSNSPTTSPAPPT-----PTTPQPPAAQSSPPPAQSSPPPVQSSPPPA---STP 328
Query: 521 SVQMFSGPLLPTPIPQPPSSSPKVSPSASPTIVSSPKISELHELPRPPTNFPSNSRLLGL 580
S P P P P S+P +P+++P S P S P PP P +
Sbjct: 329 PPAQSSPP--PVSSPPPVQSTPPPAPASTPPPASPPPFSPPPATPPPPAATPPPA----- 487
Query: 581 VGYSGPLVPRGQKVSAPNNLVISSAASPLPIPPQAMARSFSIPSSGARVTALHSSS 636
L P AP + S SP P P A+A S+ S A +L S S
Sbjct: 488 ------LTPVPATSPAPAPAKVKS-KSPAPAPAPALAPVVSLSPSEAPGPSLSSLS 634
>TC8393 weakly similar to GB|AAM61124.1|21536792|AY084557 prenylated Rab
receptor 2 {Arabidopsis thaliana;}, partial (78%)
Length = 1012
Score = 39.7 bits (91), Expect = 0.002
Identities = 35/117 (29%), Positives = 48/117 (40%)
Frame = +1
Query: 531 PTPIPQPPSSSPKVSPSASPTIVSSPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPR 590
PTP+P PP S+P + S +P+ +S L + G S P+ P
Sbjct: 130 PTPLPPPPPSAPSSTTSPNPSATAS---------------------LSAVPGRSLPIAPH 246
Query: 591 GQKVSAPNNLVISSAASPLPIPPQAMARSFSIPSSGARVTALHSSSALESSHISSIS 647
+PN +P P+PP A AR S S+ + SSS SS I S S
Sbjct: 247 -----SPNR-------NPSPMPPSASAR-ISPTSASTTTPSFRSSSPCRSSPIRSRS 378
Score = 27.7 bits (60), Expect = 6.2
Identities = 26/101 (25%), Positives = 40/101 (38%), Gaps = 5/101 (4%)
Frame = +1
Query: 572 PSNSRLLGLVGYSGPLVPRGQKVSAPNNLVI-----SSAASPLPIPPQAMARSFSIPSSG 626
P+ S L L Y+ P + S+P+ + S P P+PP + S S
Sbjct: 7 PTPSHFLSLNRYNKQPCPPPRHPSSPSPTPLRHPPPSHLPDPTPLPPPPPSAPSSTTSPN 186
Query: 627 ARVTALHSSSALESSHISSISEDIASPPLTPIALSNSRPSS 667
TA S+ S I+ S + P+ P A + P+S
Sbjct: 187 PSATASLSAVPGRSLPIAPHSPNRNPSPMPPSASARISPTS 309
>TC11087 weakly similar to UP|Q8GS92 (Q8GS92) OJ1513_F02.5 protein
(OJ1118_G09.13 protein), partial (6%)
Length = 689
Score = 39.3 bits (90), Expect = 0.002
Identities = 39/160 (24%), Positives = 66/160 (40%), Gaps = 2/160 (1%)
Frame = +1
Query: 507 LTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKVSPSASPTIVSSPKISELHELPR 566
+ NP T + + +S P TP P P SSP++ P+++P P+
Sbjct: 1 INQNPSSTCSKTTTPMATWSTPSPSTPTPSKP-SSPQIPPTSTP------------GSPK 141
Query: 567 PPTNFPSNSRLLGLVGYSGPLVPRGQKV--SAPNNLVISSAASPLPIPPQAMARSFSIPS 624
PP +NS + SG P+ + S +N ++AAS + + +PS
Sbjct: 142 PPATATTNSPSSSVSTSSGAPTPKPSRTTPSPSSNSASTTAASSSKSSTHPPSLTLCLPS 321
Query: 625 SGARVTALHSSSALESSHISSISEDIASPPLTPIALSNSR 664
S T L + S ++SIS P P+ +++R
Sbjct: 322 SPTPNTRL*AL----GSRVTSISFSRTGPSRWPMPSTSAR 429
>TC17551 weakly similar to UP|AAR24662 (AAR24662) At5g26960, partial (22%)
Length = 505
Score = 39.3 bits (90), Expect = 0.002
Identities = 40/124 (32%), Positives = 52/124 (41%), Gaps = 14/124 (11%)
Frame = +3
Query: 475 LPPPLVDGLLSS-------NHDYVSAYSKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSG 527
LPPP L +S +H S S A GP +S P PT P S + +
Sbjct: 117 LPPPFPPSLTTSFSTSSPASHPLPSLPSPS---SAAVGPTSSTP-PTSPTSAAAATSSAT 284
Query: 528 PLLPTP-----IPQPPSSSPK-VSPSASPTIVSSPKISELHELPRPPT-NFPSNSRLLGL 580
P P+P P PPSS+P PS+SP SP RP T + P+ + L
Sbjct: 285 PPSPSPPPTSASPPPPSSTPNGTPPSSSPATTPSPS--------RPSTPSSPTRASLPSA 440
Query: 581 VGYS 584
G+S
Sbjct: 441 PGFS 452
>TC8136 similar to GB|AAB60729.1|2160166|F21M12 F21M12.13 {Arabidopsis
thaliana;}, partial (70%)
Length = 1659
Score = 38.1 bits (87), Expect = 0.005
Identities = 44/165 (26%), Positives = 72/165 (42%), Gaps = 3/165 (1%)
Frame = +2
Query: 454 PTVRNA-QSVLKESNSNTATTRLPPPLVDGLLSSNHDYVSAYSKKIKRHAFSGPLTSNPM 512
P++ NA S ++S+ T + PP ++ HD +++ KR + P +P
Sbjct: 182 PSMANAPHSTHQKSHGTTVSWTWPPR------TTQHDSLTSPPSPPKRPS---PPHQSP- 331
Query: 513 PTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKVSPSASPTIVSSPKISELHELPRPPTNFP 572
P RP + + L +PP+SS S + +PT +SS + P PP +FP
Sbjct: 332 PARPST*ATT-------LSVSRSEPPASSSSWSSTPAPTRLSS-RAPAAPAAPPPPLHFP 487
Query: 573 SNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLP--IPPQA 615
+P + +AP++ SAASP P PP+A
Sbjct: 488 PKH------------LPHTARSTAPSHCAARSAASPAPPRAPPRA 586
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.313 0.129 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,652,547
Number of Sequences: 28460
Number of extensions: 200925
Number of successful extensions: 3804
Number of sequences better than 10.0: 400
Number of HSP's better than 10.0 without gapping: 2712
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3347
length of query: 669
length of database: 4,897,600
effective HSP length: 96
effective length of query: 573
effective length of database: 2,165,440
effective search space: 1240797120
effective search space used: 1240797120
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Lotus: description of TM0117b.11


