

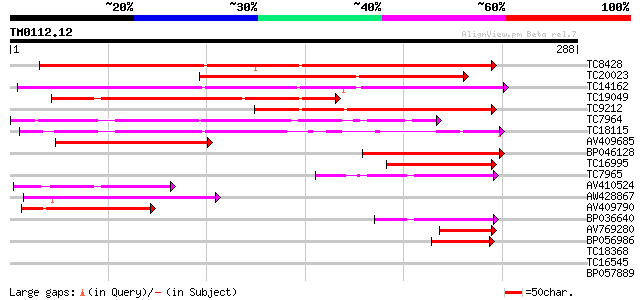
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0112.12
(288 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8428 similar to UP|Q9SNY0 (Q9SNY0) SCUTL2, partial (87%) 236 4e-63
TC20023 similar to GB|AAP13435.1|30023804|BT006327 At1g20030 {Ar... 197 1e-51
TC14162 similar to UP|OLP1_LYCES (Q41350) Osmotin-like protein p... 181 2e-46
TC19049 similar to PIR|T05694|T05694 pathogenesis-related protei... 162 5e-41
TC9212 similar to UP|TLPH_ARATH (P50699) Thaumatin-like protein ... 147 3e-36
TC7964 homologue to UP|O81926 (O81926) Thaumatin-like protein PR... 130 2e-31
TC18115 weakly similar to UP|Q93XD4 (Q93XD4) Thaumatin-like prot... 119 8e-28
AV409685 108 1e-24
BP046128 93 5e-20
TC16995 similar to PIR|T05493|T05493 pathogenesis-related protei... 91 2e-19
TC7965 homologue to UP|O81926 (O81926) Thaumatin-like protein PR... 78 2e-15
AV410524 76 6e-15
AW428867 71 2e-13
AV409790 65 2e-11
BP036640 63 5e-11
AV769280 54 4e-08
BP056986 52 9e-08
TC18368 similar to GB|AAP13435.1|30023804|BT006327 At1g20030 {Ar... 30 0.48
TC16545 28 2.4
BP057889 27 3.1
>TC8428 similar to UP|Q9SNY0 (Q9SNY0) SCUTL2, partial (87%)
Length = 1131
Score = 236 bits (601), Expect = 4e-63
Identities = 120/235 (51%), Positives = 145/235 (61%), Gaps = 3/235 (1%)
Frame = +1
Query: 16 LSFLSEVQSAS-FEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSRTLHFPKAWSGR 74
L L V SA+ F + N C +T+WPG LSG + L GF+L G S + P WSGR
Sbjct: 100 LLLLGNVASATLFTLENHCTYTVWPGTLSGNGAAVLGDGGFSLAPGASVQVTAPSGWSGR 279
Query: 75 IWARTLCGHDRDGKFSCVTADCGSDKVECIGGAKPPATLAEFTLNGADG--LDFYDVSLV 132
WART C D G +C T DC ++C GG PP TLAEFT+ A DFYDVSLV
Sbjct: 280 FWARTGCNFDGTGNGNCATGDCAGG-LKCTGGGVPPVTLAEFTIGTASNGNKDFYDVSLV 456
Query: 133 DGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGSVACMSACEAFGDP 192
DGYN+ M + A GT G C GC+ DLNG CP +L+V N TG VAC SAC AF
Sbjct: 457 DGYNVGMGVRATGGT-GDCQYAGCVADLNGSCPAELQVKGENSTGEVVACKSACAAFNTT 633
Query: 193 RYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCASANYLIIFCP 247
+CC+ ++TP TC+P+ YS FK ACP AYSYAYDD TST TC+ ++Y I FCP
Sbjct: 634 EFCCTGEHATPATCSPTHYSEIFKSACPTAYSYAYDDATSTCTCSGSDYSITFCP 798
>TC20023 similar to GB|AAP13435.1|30023804|BT006327 At1g20030 {Arabidopsis
thaliana;}, partial (46%)
Length = 418
Score = 197 bits (502), Expect = 1e-51
Identities = 93/138 (67%), Positives = 110/138 (79%), Gaps = 1/138 (0%)
Frame = +3
Query: 97 GSDKVECIG-GAKPPATLAEFTLNGADGLDFYDVSLVDGYNLPMLIVARRGTRGGCSPTG 155
GS K+EC G GA PPATLAEFTL+G+DGLDF+DVSLVDGYN+PML+V G+ C+ TG
Sbjct: 6 GSGKLECAGSGAAPPATLAEFTLDGSDGLDFFDVSLVDGYNVPMLVVPEGGSGNNCTATG 185
Query: 156 CLVDLNGGCPRDLKVARVNGTGGSVACMSACEAFGDPRYCCSEAYSTPDTCAPSVYSLFF 215
C+ DLNG CP +L+V V+G +VAC SACEAFG P+YCCS AY +P+TC PS YS F
Sbjct: 186 CVGDLNGACPSELRVTSVDGK-RNVACKSACEAFGSPQYCCSGAYGSPNTCKPSTYSQVF 362
Query: 216 KHACPRAYSYAYDDKTST 233
K ACPRAYSYAYDDK+ST
Sbjct: 363 KSACPRAYSYAYDDKSST 416
>TC14162 similar to UP|OLP1_LYCES (Q41350) Osmotin-like protein precursor,
partial (87%)
Length = 1232
Score = 181 bits (458), Expect = 2e-46
Identities = 101/255 (39%), Positives = 139/255 (53%), Gaps = 6/255 (2%)
Frame = +1
Query: 5 LLFLILLSLFTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSRT 64
LL L L + L+ + +VN C++T+WPG+ A P L GF L +
Sbjct: 49 LLISCFLILTITTHLTPSHALFLTLVNNCKYTVWPGIQPNAGHPVLAGGGFVLNPFTHLS 228
Query: 65 LHFPKA-WSGRIWARTLCGHDRDGKFSCVTADCGSDKVECIG-GAKPPATLAEFTLN-GA 121
+ P WSGR+WART C + +F+C + DCG +++C G G PATLA+F ++ G
Sbjct: 229 IPVPDTHWSGRVWARTGCTYAAGNRFTCASGDCGG-RLQCAGAGGSAPATLAQFDMHHGN 405
Query: 122 DGLDFYDVSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDL--KVARVNGTGGS 179
Y VSLVDG+N+PM + G +G C GC DL CP L +V V+G
Sbjct: 406 ADFSSYSVSLVDGFNVPMTVTPHEG-KGVCPVVGCRSDLLATCPPVLHHRVPAVHGP--V 576
Query: 180 VACMSACEAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCASA 239
VAC S CEAF CC ++ TC PSVYS FFKHACP +++A+D + T+ C+S
Sbjct: 577 VACKSGCEAFHSDELCCRNHFNNAQTCKPSVYSTFFKHACPATFTFAHDTPSLTHECSSP 756
Query: 240 NYL-IIFCPLPYTRY 253
L +IFC TR+
Sbjct: 757 RELKVIFCH*KRTRF 801
>TC19049 similar to PIR|T05694|T05694 pathogenesis-related protein
F20M13.220 - Arabidopsis thaliana
{Arabidopsis thaliana;}, partial (36%)
Length = 861
Score = 162 bits (411), Expect = 5e-41
Identities = 81/148 (54%), Positives = 97/148 (64%), Gaps = 1/148 (0%)
Frame = +3
Query: 22 VQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSRTLHFPKAWSGRIWARTLC 81
V A+F VNKC T+WPG+ P + +TGF L G S++ P WSGR WART C
Sbjct: 432 VSGATFSFVNKCDFTVWPGIFG---KPEIGSTGFELTKGSSKSFQAPTGWSGRFWARTGC 602
Query: 82 GHDRDGKFSCVTADCGSDKVECIGG-AKPPATLAEFTLNGADGLDFYDVSLVDGYNLPML 140
D G+ +C TADCGS ++ C G A PPATLAEFTL G +DFYDVSLVDGYNLPM+
Sbjct: 603 QFDSSGRGTCATADCGSGEINCNGATALPPATLAEFTL-GTGSMDFYDVSLVDGYNLPMM 779
Query: 141 IVARRGTRGGCSPTGCLVDLNGGCPRDL 168
+VA G+ G CS TGC DLN CP +L
Sbjct: 780 VVASGGS-GSCSATGCGADLNRKCPSEL 860
>TC9212 similar to UP|TLPH_ARATH (P50699) Thaumatin-like protein precursor,
partial (51%)
Length = 662
Score = 147 bits (370), Expect = 3e-36
Identities = 72/123 (58%), Positives = 84/123 (67%)
Frame = +1
Query: 125 DFYDVSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGSVACMS 184
DFYDVSLVDGYNLP+ I +G+ G CS GC+ DLN CP L+V R VAC S
Sbjct: 1 DFYDVSLVDGYNLPISITPLKGS-GKCSYAGCVRDLNIMCPVGLQV-RSRDNKRVVACKS 174
Query: 185 ACEAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCASANYLII 244
AC AF PRYCC+ +Y +P +C P+ YS FK ACP+AYSYAYDD TS TC A+YLI
Sbjct: 175 ACAAFNSPRYCCTGSYGSPQSCKPTAYSRIFKTACPKAYSYAYDDPTSISTCTHASYLIT 354
Query: 245 FCP 247
FCP
Sbjct: 355 FCP 363
>TC7964 homologue to UP|O81926 (O81926) Thaumatin-like protein PR-5b
precursor (Fragment), partial (74%)
Length = 778
Score = 130 bits (328), Expect = 2e-31
Identities = 82/220 (37%), Positives = 109/220 (49%), Gaps = 1/220 (0%)
Frame = +2
Query: 1 MGRLLLFLILLSLFTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESG 60
MG L L +LL L T S ++ ++A+FEIVN C +T+W G G L+ G
Sbjct: 35 MGYLTLSSLLLML-TASLVTLTRAANFEIVNNCPYTVWAAASPGG--------GRRLDRG 187
Query: 61 KSRTLHFPKAWS-GRIWARTLCGHDRDGKFSCVTADCGSDKVECIGGAKPPATLAEFTLN 119
++ L + RIW RT C D G+ C T DC + + C G PP TLAEF LN
Sbjct: 188 QTWNLGVNAGTAMARIWGRTGCNFDGAGRGRCQTGDC-TGGLNCQGWGVPPNTLAEFALN 364
Query: 120 GADGLDFYDVSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGS 179
DFYD+SLVDG+N+PM GGC C D+NG CP +L+ GG
Sbjct: 365 QYGNQDFYDISLVDGFNIPMDFFP---LNGGCHKISCTADINGQCPNELR-----APGG- 517
Query: 180 VACMSACEAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHAC 219
C + C A+ +CC+ TC P+ + FFK C
Sbjct: 518 --CNNPCTAYKTNEFCCTNGQG---TCGPN*FLRFFKDRC 622
>TC18115 weakly similar to UP|Q93XD4 (Q93XD4) Thaumatin-like protein,
partial (63%)
Length = 792
Score = 119 bits (297), Expect = 8e-28
Identities = 83/251 (33%), Positives = 112/251 (44%), Gaps = 5/251 (1%)
Frame = +2
Query: 6 LFLILLSLFTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSRTL 65
LF ++ FTL+ Q+A F+I N+C + +WP L G G L SG++ L
Sbjct: 26 LFSLIALCFTLA-----QAARFDITNRCSYPVWPASLPGG--------GRRLNSGETWPL 166
Query: 66 HFPKAWSGRIWARTLCGHDRDGKFSCVTADCGSDKVECIGGAKPPATLAEFTLNGADGLD 125
+ SGRIW RT C D G+ SC T DCG + C +PP TLAEFTLNG + D
Sbjct: 167 NLNAGSSGRIWGRTGCNFDGSGRGSCQTGDCGG-ALSCSLSGRPPTTLAEFTLNGGNNQD 343
Query: 126 FYDVSLVDGYNLPMLIVARRGTRGGCSPTGCLVDLNGGCPRDLKVARVNGTGGSVACMSA 185
+YD+S++DG+N+PM SPT + GC R L+
Sbjct: 344 YYDISVIDGFNVPMEF----------SPT------SNGCTRSLR---------------- 427
Query: 186 CEAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTC-ASANYLII 244
C A CP AY Y DD T T++C +Y ++
Sbjct: 428 CHA----------------------------DKCPDAYLYPSDD-TKTHSCPGGTHYRVV 520
Query: 245 FCP----LPYT 251
FCP LP+T
Sbjct: 521 FCP*SWTLPFT 553
>AV409685
Length = 270
Score = 108 bits (270), Expect = 1e-24
Identities = 44/80 (55%), Positives = 57/80 (71%)
Frame = +2
Query: 24 SASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSRTLHFPKAWSGRIWARTLCGH 83
S +F +VNKC +T+WPG+L+ A PLP TGF L++G+S+ + P W GR W RTLC
Sbjct: 29 SRTFTVVNKCDYTVWPGILTNAGVTPLPITGFVLQTGESKLIPAPANWGGRFWGRTLCSQ 208
Query: 84 DRDGKFSCVTADCGSDKVEC 103
D GKF+CVT DCGS K+EC
Sbjct: 209 DSAGKFTCVTGDCGSGKLEC 268
>BP046128
Length = 514
Score = 93.2 bits (230), Expect = 5e-20
Identities = 38/72 (52%), Positives = 51/72 (70%)
Frame = -1
Query: 180 VACMSACEAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCASA 239
V C SAC AF + +YCC+ + +P+ C PS Y+LFFK CP AYSY DD++ST++C+ A
Sbjct: 511 VGCNSACVAFNEAKYCCTGEFESPELCKPSEYALFFKKKCPLAYSYPSDDESSTFSCSRA 332
Query: 240 NYLIIFCPLPYT 251
+Y I FCP YT
Sbjct: 331 DYTITFCP*TYT 296
>TC16995 similar to PIR|T05493|T05493 pathogenesis-related protein 19K4.140
- Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(22%)
Length = 614
Score = 90.9 bits (224), Expect = 2e-19
Identities = 37/56 (66%), Positives = 43/56 (76%)
Frame = +1
Query: 192 PRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTCASANYLIIFCP 247
P YCC+ A+S P TC PSVYS FK ACP +YSYAYDD TST+TC+ A+Y I FCP
Sbjct: 1 PEYCCNGAFSNPSTCKPSVYSEMFKSACPESYSYAYDDATSTFTCSGADYTITFCP 168
>TC7965 homologue to UP|O81926 (O81926) Thaumatin-like protein PR-5b
precursor (Fragment), partial (37%)
Length = 633
Score = 77.8 bits (190), Expect = 2e-15
Identities = 38/94 (40%), Positives = 52/94 (54%), Gaps = 1/94 (1%)
Frame = +2
Query: 156 CLVDLNGGCPRDLKVARVNGTGGSVACMSACEAFGDPRYCCSEAYSTPDTCAPSVYSLFF 215
C D+NG CP +L+ GG C + C + +CC+ T C P+ +S FF
Sbjct: 8 CTADINGQCPNELRAP-----GG---CNNPCTVYKTNEFCCTNGQGT---CGPTNFSRFF 154
Query: 216 KHACPRAYSYAYDDKTSTYTC-ASANYLIIFCPL 248
K C +YSY DD TST+TC A +NY ++FCPL
Sbjct: 155 KDRCHDSYSYPQDDPTSTFTCPAGSNYKVVFCPL 256
>AV410524
Length = 402
Score = 76.3 bits (186), Expect = 6e-15
Identities = 36/82 (43%), Positives = 49/82 (58%)
Frame = +3
Query: 3 RLLLFLILLSLFTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKS 62
R ++FL++L+ + + +F VNKC HT+WPG+L P L TTGF L+ G +
Sbjct: 174 RFIIFLLILAFKGV----RISGTTFTFVNKCDHTVWPGILG---KPDLETTGFELKRGNT 332
Query: 63 RTLHFPKAWSGRIWARTLCGHD 84
RT+ P WSGR WART C D
Sbjct: 333 RTIPAPAGWSGRFWARTGCKFD 398
>AW428867
Length = 375
Score = 71.2 bits (173), Expect = 2e-13
Identities = 36/102 (35%), Positives = 54/102 (52%), Gaps = 2/102 (1%)
Frame = +1
Query: 8 LILLSLFTLSFLS--EVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSRTL 65
++ L+LF +S +S +V +A+ NKC H +WPG+ G P L GF L K+ +L
Sbjct: 70 ILFLTLFLISHISVLKVSAAAAIFYNKCPHPVWPGIQPGPGKPVLARGGFKLLPNKAYSL 249
Query: 66 HFPKAWSGRIWARTLCGHDRDGKFSCVTADCGSDKVECIGGA 107
P WSGR W R C D G+ + A + ++ +GGA
Sbjct: 250 QLPALWSGRFWGRHDCTFDASGRGTAPPATAELNLLQRLGGA 375
>AV409790
Length = 421
Score = 64.7 bits (156), Expect = 2e-11
Identities = 29/68 (42%), Positives = 45/68 (65%)
Frame = +2
Query: 7 FLILLSLFTLSFLSEVQSASFEIVNKCRHTIWPGLLSGANSPPLPTTGFALESGKSRTLH 66
+ IL++L L+ + V++ +F +N+C +T+WPG+L+ A S PL +TGF L + SRT
Sbjct: 218 YFILITLLLLT-PTGVRATTFTFLNRCDYTVWPGILANAGSSPLESTGFELPTATSRTFQ 394
Query: 67 FPKAWSGR 74
P WSGR
Sbjct: 395 APTGWSGR 418
>BP036640
Length = 468
Score = 63.2 bits (152), Expect = 5e-11
Identities = 28/64 (43%), Positives = 38/64 (58%), Gaps = 1/64 (1%)
Frame = -2
Query: 186 CEAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYSYAYDDKTSTYTC-ASANYLII 244
C + +CC+ T C P+ +S FFK C +YSY DD TST+TC A +NY ++
Sbjct: 464 CTVYKTNEFCCTNGQGT---CGPTNFSRFFKDRCHDSYSYPQDDPTSTFTCPAGSNYKVV 294
Query: 245 FCPL 248
FCPL
Sbjct: 293 FCPL 282
>AV769280
Length = 327
Score = 53.5 bits (127), Expect = 4e-08
Identities = 20/29 (68%), Positives = 25/29 (85%)
Frame = -3
Query: 219 CPRAYSYAYDDKTSTYTCASANYLIIFCP 247
CP++YSYAYDD TST+TC+ A+Y I FCP
Sbjct: 325 CPKSYSYAYDDATSTFTCSGADYTITFCP 239
>BP056986
Length = 450
Score = 52.4 bits (124), Expect = 9e-08
Identities = 20/32 (62%), Positives = 25/32 (77%)
Frame = -3
Query: 215 FKHACPRAYSYAYDDKTSTYTCASANYLIIFC 246
FK ACP +YSYAYDD TS +TC+ +Y+I FC
Sbjct: 445 FKEACPTSYSYAYDDPTSIFTCSGTDYVIAFC 350
>TC18368 similar to GB|AAP13435.1|30023804|BT006327 At1g20030 {Arabidopsis
thaliana;}, partial (7%)
Length = 603
Score = 30.0 bits (66), Expect = 0.48
Identities = 12/17 (70%), Positives = 13/17 (75%)
Frame = +3
Query: 235 TCASANYLIIFCPLPYT 251
TCASA+Y I FCP P T
Sbjct: 3 TCASADYTITFCPSPTT 53
>TC16545
Length = 660
Score = 27.7 bits (60), Expect = 2.4
Identities = 16/35 (45%), Positives = 22/35 (62%)
Frame = -3
Query: 4 LLLFLILLSLFTLSFLSEVQSASFEIVNKCRHTIW 38
L+LFL+ +LS +S VQS S+ + N RH IW
Sbjct: 400 LMLFLVP----SLSRVSLVQSPSYPLSNYYRHKIW 308
>BP057889
Length = 473
Score = 27.3 bits (59), Expect = 3.1
Identities = 12/43 (27%), Positives = 19/43 (43%)
Frame = -2
Query: 182 CMSACEAFGDPRYCCSEAYSTPDTCAPSVYSLFFKHACPRAYS 224
C A P+YCC + S+ +C S + C R+Y+
Sbjct: 424 CQPAVAYVRPPKYCCIRSLSSCISCTACRSSTDLRTTCRRSYT 296
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.324 0.141 0.457
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,422,692
Number of Sequences: 28460
Number of extensions: 108226
Number of successful extensions: 698
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 670
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 673
length of query: 288
length of database: 4,897,600
effective HSP length: 89
effective length of query: 199
effective length of database: 2,364,660
effective search space: 470567340
effective search space used: 470567340
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0112.12


