

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0109.3
(297 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
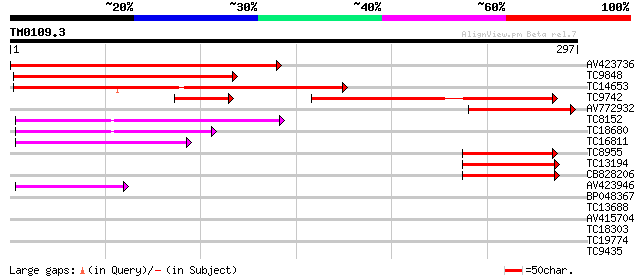
Score E
Sequences producing significant alignments: (bits) Value
AV423736 290 3e-79
TC9848 weakly similar to UP|Q8LBR0 (Q8LBR0) Phloem-specific lect... 191 1e-49
TC14653 weakly similar to UP|Q9FLU7 (Q9FLU7) Phloem-specific lec... 164 1e-41
TC9742 similar to UP|Q8LBR0 (Q8LBR0) Phloem-specific lectin PP2-... 131 1e-31
AV772932 104 2e-23
TC8152 97 2e-21
TC18680 82 1e-16
TC16811 66 8e-12
TC8955 weakly similar to UP|Q9FLU7 (Q9FLU7) Phloem-specific lect... 62 1e-10
TC13194 weakly similar to UP|Q94EW1 (Q94EW1) Nictaba, partial (29%) 57 3e-09
CB828206 48 2e-06
AV423946 48 2e-06
BP048367 38 0.002
TC13688 similar to UP|O22161 (O22161) At2g44900 protein, partial... 30 0.64
AV415704 28 1.4
TC18303 similar to GB|AAG32022.1|11141605|AF277458 LEUNIG {Arabi... 28 1.9
TC19774 weakly similar to UP|AAR92268 (AAR92268) At1g47570, part... 28 2.4
TC9435 similar to UP|Q9AXJ7 (Q9AXJ7) Phosphatase-like protein Mt... 27 4.1
>AV423736
Length = 454
Score = 290 bits (741), Expect = 3e-79
Identities = 142/142 (100%), Positives = 142/142 (100%)
Frame = +2
Query: 1 MISLPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPS 60
MISLPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPS
Sbjct: 29 MISLPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPS 208
Query: 61 ALQFSSYKQLFHALCSPLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRS 120
ALQFSSYKQLFHALCSPLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRS
Sbjct: 209 ALQFSSYKQLFHALCSPLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRS 388
Query: 121 NPESRFREVAELRTVNWLEIEG 142
NPESRFREVAELRTVNWLEIEG
Sbjct: 389 NPESRFREVAELRTVNWLEIEG 454
>TC9848 weakly similar to UP|Q8LBR0 (Q8LBR0) Phloem-specific lectin
PP2-like protein, partial (19%)
Length = 394
Score = 191 bits (486), Expect = 1e-49
Identities = 91/118 (77%), Positives = 106/118 (89%), Gaps = 1/118 (0%)
Frame = +1
Query: 3 SLPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPSAL 62
+LPE+CVSEILSHTSPPDACRFS++SSTLRSAA+SDM+WRSFLPSDY DI+SRA+NP L
Sbjct: 37 TLPEECVSEILSHTSPPDACRFSMLSSTLRSAANSDMLWRSFLPSDYSDIISRALNPLFL 216
Query: 63 -QFSSYKQLFHALCSPLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWR 119
SS+K LF ALC+PLLLDGG FKLDK SGKKSYILSAR+LSITWSSDP++W+W+
Sbjct: 217 NSSSSFKDLFKALCNPLLLDGGTMIFKLDKSSGKKSYILSARELSITWSSDPLYWTWK 390
>TC14653 weakly similar to UP|Q9FLU7 (Q9FLU7) Phloem-specific lectin-like
protein, partial (45%)
Length = 643
Score = 164 bits (416), Expect = 1e-41
Identities = 91/180 (50%), Positives = 114/180 (62%), Gaps = 5/180 (2%)
Frame = +2
Query: 3 SLPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSR----AVN 58
+LPE C++ ILS TSP D R +LVSS RSAADSD VW FLP D+ I+S A
Sbjct: 110 NLPEGCIANILSLTSPLDVGRLALVSSAFRSAADSDAVWDRFLPDDFNSIISHSSSTAAA 289
Query: 59 PSALQFSSYKQLFHALC-SPLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWS 117
S+ F S K L+ +L +PL++ G KSF+L ++GKK Y+LSAR LSI W P +W
Sbjct: 290 DSSSSFKSKKDLYLSLSHNPLIIGDGKKSFQL--VNGKKCYMLSARALSIVWGDTPRYWR 463
Query: 118 WRSNPESRFREVAELRTVNWLEIEGKIRTGVLTPNTSYGAYLIMKVSHRGYGLDSAPAEV 177
W S PE+RF EVAEL +V WLEI G I +L+P T YGAYL+ K S YG + EV
Sbjct: 464 WISLPEARFSEVAELVSVCWLEIRGWINNKMLSPETMYGAYLVFKPSGGAYGFEFQSVEV 643
>TC9742 similar to UP|Q8LBR0 (Q8LBR0) Phloem-specific lectin PP2-like
protein, partial (20%)
Length = 794
Score = 131 bits (330), Expect = 1e-31
Identities = 74/131 (56%), Positives = 87/131 (65%), Gaps = 2/131 (1%)
Frame = +1
Query: 159 LIMKVSHRGYGLDSAPAEVSVAVGGRVLRRGKAYLGQKDEKKV-EMETLFYGNRRDIFRN 217
LIM VSHRGYGLDSAP+EVS V V+ K YL K+EK + E TLF G RD+ R
Sbjct: 235 LIMNVSHRGYGLDSAPSEVSSTVANNVMHTKKVYLWNKNEKNMCEKYTLFRGFHRDVARE 414
Query: 218 RVIQLEQLEEEGKDGGVIPVPGKRDDGWMEIELGEFFSGGG-DVEIKMGLREVGYQLKGG 276
+I +Q+ P KRDDGWMEIE+GEFF G D E+K+ + EVGYQLKGG
Sbjct: 415 TLIIQDQINSG---------PLKRDDGWMEIEVGEFFCDGEIDEEVKVSVMEVGYQLKGG 567
Query: 277 LVVEGIEVRPK 287
L+VEGIEVRPK
Sbjct: 568 LIVEGIEVRPK 600
Score = 60.1 bits (144), Expect = 4e-10
Identities = 26/31 (83%), Positives = 30/31 (95%)
Frame = +2
Query: 87 FKLDKLSGKKSYILSARDLSITWSSDPMFWS 117
FKLDK SGKKSYILSAR+LSITWSSDP++W+
Sbjct: 143 FKLDKSSGKKSYILSARELSITWSSDPLYWT 235
>AV772932
Length = 481
Score = 104 bits (259), Expect = 2e-23
Identities = 50/56 (89%), Positives = 51/56 (90%)
Frame = -3
Query: 241 RDDGWMEIELGEFFSGGGDVEIKMGLREVGYQLKGGLVVEGIEVRPKQTQSTQKTK 296
RDDGWMEIELGEFFSGGGDVEIKMGLRE+GYQLKGGLV EGIEVRPK TQST K
Sbjct: 479 RDDGWMEIELGEFFSGGGDVEIKMGLREMGYQLKGGLVFEGIEVRPKPTQSTPTPK 312
>TC8152
Length = 715
Score = 97.4 bits (241), Expect = 2e-21
Identities = 54/141 (38%), Positives = 77/141 (54%)
Frame = +1
Query: 4 LPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPSALQ 63
LPE CV+ IL + PP C+ + ++ R+A+ +D VW S LP++Y IV R +
Sbjct: 199 LPESCVALILGYADPPQICQLATLNRAFRAASSADFVWESKLPANYRAIV-RKIFTDFPS 375
Query: 64 FSSYKQLFHALCSPLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRSNPE 123
SS + + ALC LDGG K LDK +GK LS + LSIT D +W E
Sbjct: 376 DSSKRDTYAALCRLNTLDGGTKKAWLDKSTGKICMCLSFKGLSITGIDDRRYWKDIHTEE 555
Query: 124 SRFREVAELRTVNWLEIEGKI 144
SRF VA L+ W +++G++
Sbjct: 556 SRFGTVAYLQQTWWFQVDGEV 618
>TC18680
Length = 492
Score = 82.0 bits (201), Expect = 1e-16
Identities = 40/105 (38%), Positives = 63/105 (59%)
Frame = +1
Query: 4 LPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPSALQ 63
+PE+CV+ + H +PP+ C + ++ R AA +D VW + LPS+Y+D++ A+ P Q
Sbjct: 181 IPENCVARVFLHLTPPEICNLARLNRAFRGAASADSVWETKLPSNYKDLL-HALPPERFQ 357
Query: 64 FSSYKQLFHALCSPLLLDGGNKSFKLDKLSGKKSYILSARDLSIT 108
S K +F L P D GNK LD+++G+ +SA+ LSIT
Sbjct: 358 NLSKKDIFALLSRPQPFDDGNKEVWLDRVTGRVCMSISAKALSIT 492
>TC16811
Length = 509
Score = 65.9 bits (159), Expect = 8e-12
Identities = 28/92 (30%), Positives = 50/92 (53%)
Frame = +2
Query: 4 LPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPSALQ 63
+PE+C+S +L P + C+ + V+ A+ +D +W S LP Y+ +V++ + L
Sbjct: 233 VPENCISSLLMSLDPQEICKLARVNKAFHRASSADFLWESKLPPGYKFLVNKVLGEEKLG 412
Query: 64 FSSYKQLFHALCSPLLLDGGNKSFKLDKLSGK 95
+ K+++ LC P DGG K LD+ S +
Sbjct: 413 SMTKKEIYAKLCQPNFFDGGAKEIWLDRCSAQ 508
>TC8955 weakly similar to UP|Q9FLU7 (Q9FLU7) Phloem-specific lectin-like
protein, partial (12%)
Length = 693
Score = 62.0 bits (149), Expect = 1e-10
Identities = 32/52 (61%), Positives = 40/52 (76%), Gaps = 2/52 (3%)
Frame = +2
Query: 238 PGKRDDGWMEIELGEFF-SGGGDVEIKMGLREV-GYQLKGGLVVEGIEVRPK 287
P +R DGW+E+ELGEFF GG D E++M + EV G KGGLVV+GIE+RPK
Sbjct: 41 PKERADGWLEMELGEFFCEGGEDREVEMAVCEVKGGDWKGGLVVQGIEIRPK 196
>TC13194 weakly similar to UP|Q94EW1 (Q94EW1) Nictaba, partial (29%)
Length = 471
Score = 57.4 bits (137), Expect = 3e-09
Identities = 28/53 (52%), Positives = 38/53 (70%), Gaps = 2/53 (3%)
Frame = +1
Query: 238 PGKRDDGWMEIELGEFFSGG-GDVEIKMGLREV-GYQLKGGLVVEGIEVRPKQ 288
P R DGW+EIE+GEF + D E++M + E+ GY K GL++EGIEVRPK+
Sbjct: 28 PAVRSDGWLEIEMGEFSNSSLEDEELQMSVIEIQGYSSKSGLILEGIEVRPKE 186
>CB828206
Length = 466
Score = 48.1 bits (113), Expect = 2e-06
Identities = 26/53 (49%), Positives = 37/53 (69%), Gaps = 2/53 (3%)
Frame = +1
Query: 238 PGKRDDGWMEIELGEFFSGG-GDVEIKMGLREVGYQ-LKGGLVVEGIEVRPKQ 288
P R DG +EIE+GEFF+ G D E+++ + EV + K GL +EGIEVRP++
Sbjct: 49 PSVRSDG*LEIEMGEFFNSGLEDEELQLSILEVNSRNWKSGLHLEGIEVRPRE 207
>AV423946
Length = 426
Score = 47.8 bits (112), Expect = 2e-06
Identities = 18/59 (30%), Positives = 32/59 (53%)
Frame = +1
Query: 4 LPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPSAL 62
+PE C+S + + PPD C+ + V+ A+ +D VW S LP Y+ + ++ + L
Sbjct: 241 IPESCISSLFMNLDPPDICKLARVNRAFHRASSADFVWESKLPPSYKFLANKILGEENL 417
>BP048367
Length = 205
Score = 37.7 bits (86), Expect = 0.002
Identities = 15/19 (78%), Positives = 18/19 (93%)
Frame = -1
Query: 269 VGYQLKGGLVVEGIEVRPK 287
VGY LKGGL++EG+EVRPK
Sbjct: 205 VGYPLKGGLIIEGLEVRPK 149
>TC13688 similar to UP|O22161 (O22161) At2g44900 protein, partial (7%)
Length = 621
Score = 29.6 bits (65), Expect = 0.64
Identities = 20/57 (35%), Positives = 26/57 (45%)
Frame = -3
Query: 4 LPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPS 60
LP+D V ++LS S D FS T RS S +W S D+ S +PS
Sbjct: 154 LPDDTVIQLLSCLSYRDRASFSSSCRTWRSLGSSPCLWTSL------DLRSHKFDPS 2
>AV415704
Length = 355
Score = 28.5 bits (62), Expect = 1.4
Identities = 19/66 (28%), Positives = 29/66 (43%)
Frame = +1
Query: 1 MISLPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPS 60
+ SLP+D +I S D C S R SD +W S + + + + A+ PS
Sbjct: 1 LASLPQDVTFKITSLLQVRDLCALGCCSRFCRDLCFSDCIWESLVRTRWPLL---ALAPS 171
Query: 61 ALQFSS 66
+ SS
Sbjct: 172SSSSSS 189
>TC18303 similar to GB|AAG32022.1|11141605|AF277458 LEUNIG {Arabidopsis
thaliana;} , partial (12%)
Length = 571
Score = 28.1 bits (61), Expect = 1.9
Identities = 23/80 (28%), Positives = 36/80 (44%), Gaps = 2/80 (2%)
Frame = +1
Query: 36 DSDMVWRSFLPSDYEDIVSRAVNPSALQFSSYKQLFHALCSPLLLDGGNKSFKLDKLSGK 95
DS VW SD E + + N S + +FH LL+ G +S +L ++
Sbjct: 58 DSVRVWNLGTGSDGECVHELSCNGSKF----HSCVFHPTYPSLLVIGCYQSMELWNIAEN 225
Query: 96 KSYILSARD--LSITWSSDP 113
K+ LSA D +++ W P
Sbjct: 226 KTMTLSAHDGLITLPWLFQP 285
>TC19774 weakly similar to UP|AAR92268 (AAR92268) At1g47570, partial (19%)
Length = 642
Score = 27.7 bits (60), Expect = 2.4
Identities = 14/28 (50%), Positives = 16/28 (57%), Gaps = 2/28 (7%)
Frame = +2
Query: 28 SSTLRSAADSDMVWRSFLPSD--YEDIV 53
SS SA SD +W +PSD Y DIV
Sbjct: 137 SSASASAIPSDQIWAKLVPSDSRYSDIV 220
>TC9435 similar to UP|Q9AXJ7 (Q9AXJ7) Phosphatase-like protein Mtc923,
partial (17%)
Length = 486
Score = 26.9 bits (58), Expect = 4.1
Identities = 22/61 (36%), Positives = 28/61 (45%), Gaps = 7/61 (11%)
Frame = -3
Query: 229 GKDGGVIPVPGKRDDGWMEIEL-------GEFFSGGGDVEIKMGLREVGYQLKGGLVVEG 281
G D GV+ V + DG +E+E+ G F GGGD L E G + G V G
Sbjct: 175 GVD*GVVVVVEEGGDGGVEVEIVAFEEEEGLLFGGGGD------LAECGDECGGVAVGFG 14
Query: 282 I 282
I
Sbjct: 13 I 11
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,101,609
Number of Sequences: 28460
Number of extensions: 69460
Number of successful extensions: 429
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 419
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 419
length of query: 297
length of database: 4,897,600
effective HSP length: 90
effective length of query: 207
effective length of database: 2,336,200
effective search space: 483593400
effective search space used: 483593400
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 55 (25.8 bits)
Lotus: description of TM0109.3


