

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
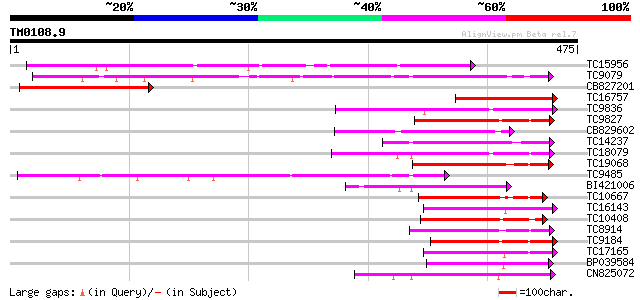
Query= TM0108.9
(475 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC15956 weakly similar to UP|LGT_CITUN (Q9MB73) Limonoid UDP-glu... 214 3e-56
TC9079 weakly similar to UP|Q8W4G1 (Q8W4G1) UDP-glucose glucosyl... 164 2e-41
CB827201 143 7e-35
TC16757 weakly similar to UP|Q9SBQ2 (Q9SBQ2) Anthocyanin 5-O-glu... 125 1e-29
TC9836 weakly similar to UP|Q9LXV0 (Q9LXV0) Glucosyltransferase-... 117 4e-27
TC9827 weakly similar to UP|Q8S9A2 (Q8S9A2) Glucosyltransferase-... 115 2e-26
CB829602 110 4e-25
TC14237 weakly similar to UP|AAR06913 (AAR06913) UDP-glycosyltra... 110 5e-25
TC18079 weakly similar to UP|UFO5_MANES (Q40287) Flavonol 3-O-gl... 106 9e-24
TC19068 weakly similar to UP|AAR06913 (AAR06913) UDP-glycosyltra... 97 7e-21
TC9485 similar to UP|Q8S999 (Q8S999) Glucosyltransferase-10, par... 96 1e-20
BI421006 92 2e-19
TC10667 similar to UP|Q9M051 (Q9M051) Glucuronosyl transferase-l... 89 2e-18
TC16143 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltrans... 87 5e-18
TC10408 weakly similar to UP|Q9FUJ6 (Q9FUJ6) UDP-glucosyltransfe... 86 1e-17
TC8914 similar to UP|Q9ZWS2 (Q9ZWS2) Flavonoid 3-O-galactosyl tr... 86 1e-17
TC9184 weakly similar to UP|HQGT_RAUSE (Q9AR73) Hydroquinone glu... 84 7e-17
TC17165 similar to UP|Q9SMG6 (Q9SMG6) Betanidin-5-O-glucosyltran... 82 2e-16
BP039584 81 4e-16
CN825072 81 4e-16
>TC15956 weakly similar to UP|LGT_CITUN (Q9MB73) Limonoid
UDP-glucosyltransferase (Limonoid glucosyltransferase)
(Limonoid GTase) (LGTase) , partial (65%)
Length = 1169
Score = 214 bits (544), Expect = 3e-56
Identities = 140/388 (36%), Positives = 206/388 (53%), Gaps = 12/388 (3%)
Frame = +2
Query: 15 LIVSYPGQGQINPTLQFSKRLIAMGAHVTIVTTLHMLRRMTTKTTIPGLSYTAFSDG--- 71
L+VS+P QG INP L+ +K L A G+ VT+ TT + M T T I + T SDG
Sbjct: 44 LLVSFPAQGHINPLLRLAKCLAAKGSSVTLATTQTAGKSMRTATNITDKAPTPISDGSLK 223
Query: 72 ---FDDGFIVD---VATHFNLYTPELKRRGSEFITNLILFAENEGHPFTCLIHTLFVPWA 125
FDDG D + + +LY +L+ G + I +I + +C+I+ F+PW
Sbjct: 224 FEFFDDGLGPDDDSIRGNLSLYLQQLELVGRQQILKMIKEHADSNRQISCIINNPFLPWV 403
Query: 126 PQVARELNLITAKLWIEPTTVMSILYNYFLGGDYIAKKNTVESSCSIEFSGLPFSLSPHD 185
VA E + +A LWI+ + V + Y+YF + + + + ++ L +
Sbjct: 404 VDVAEEQGIPSALLWIQSSAVFTAYYSYF--HNLVTFPSKEDPLADVQLPSTSIVLKHCE 577
Query: 186 MPSYLISKGSYVL--PLFKEDFKELRG*ANTTILVNTFEALEPEAPRAVDKLNMI-PIGL 242
+P +L +Y L E FK L ILV+T+E LE + + K +I P+G
Sbjct: 578 IPDFLHPSCTYPFLGTLILEQFKNLN--KTFCILVDTYEELEHDFIDYLSKTFLIRPVGP 751
Query: 243 LIPSAFLDRKDFAHDTSFGGDIIQAPSNDYIEWLDSKPPSSVVYVSFGSYIVLSKTQMDE 302
L S+ A + GD ++ S+D IEWL+SKP SSVVYVSFGS + L + Q+DE
Sbjct: 752 LFKSSQ------AKSAAIRGDFMK--SDDCIEWLNSKPQSSVVYVSFGSIVYLPQEQVDE 907
Query: 303 IARALLDCRHPFLWVIREKEKFQDGEKEKEEELGNIEELEEVGKIAKWCSQVEVLSHRSM 362
IA L + FLWV++ + G + G +EE E G++ +W Q EVL+H S+
Sbjct: 908 IAFGLKKSQVSFLWVMKPPPEVT-GLQPHVLPYGFLEETGERGRVVQWSPQEEVLAHPSV 1084
Query: 363 GCFLTHCGWNSTMESLVSRVPMVAFPQW 390
CFLTHCGWNS+ME+L S VP++ FP W
Sbjct: 1085SCFLTHCGWNSSMEALTSGVPVLTFPAW 1168
>TC9079 weakly similar to UP|Q8W4G1 (Q8W4G1) UDP-glucose
glucosyltransferase, partial (65%)
Length = 1585
Score = 164 bits (416), Expect = 2e-41
Identities = 137/467 (29%), Positives = 216/467 (45%), Gaps = 31/467 (6%)
Frame = +1
Query: 20 PGQGQINPTLQFSKRLIAMGAHVTIVTTLHMLRRMTTKTT------IPGLSYTAFSDGFD 73
P QG INP Q +K L G H+T V T + +R+ +P + DG +
Sbjct: 40 PLQGHINPLFQLAKLLHLRGFHITFVHTEYNHKRLIKSRGPNALDGLPDFVFETIPDGLE 219
Query: 74 DGFIVDVATHFNLYT--PELKRRGSEFITNLILFAENEGH-----PFTCLIHTLFVPWAP 126
DG D +LY+ ++ + +L+ P TCL+ L + +
Sbjct: 220 DG---DGEVSQDLYSLCDSIRNKCHLPFQDLLAKLHRSASAGLVPPVTCLVSDLLMTFTI 390
Query: 127 QVARELNLITAKLW-IEPTTVMSILY---------------NYFLGGDYIAKKNTVESSC 170
Q A++L L L +T MS L+ +Y G K + +
Sbjct: 391 QAAQQLQLPILLLCPASASTFMSFLHFQTLLDKGVIPLKDESYLTNGYLDTKVDWIPGMQ 570
Query: 171 SIEFSGLPFSLSPHDMPSYLISKGSYVLPLFKEDFKELRG*ANTTILVNTFEALEPEAPR 230
+ LP + D ++ F + + R + I+ NTF LE +
Sbjct: 571 NFRLKDLPDFIRTTDPNDIMLE--------FMVEVAD-RAKRASAIVFNTFNELERDVLS 723
Query: 231 AVDKL--NMIPIGLLIPSAFLDRKDFAHDTSFGGDIIQAPSNDYIEWLDSKPPSSVVYVS 288
A+ + ++ PIG +FL++ S G ++ + + ++WL+SK P SVVYV+
Sbjct: 724 ALSTMLPSLYPIGPF--PSFLNQAPQNQLASLGSNLWKEDTK-CLQWLESKEPGSVVYVN 894
Query: 289 FGSYIVLSKTQMDEIARALLDCRHPFLWVIREKEKFQDGEKEKEEELGNIEELEEVGKIA 348
FGS V+S Q+ E A L + + PFLW+IR + DG + ++E G IA
Sbjct: 895 FGSITVMSPEQLLEFAWGLANGKKPFLWIIRP-DLVADGSAILSPKF--VDETSNRGLIA 1065
Query: 349 KWCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQNTNAKLIEDVWKIGV 408
WC Q +VL+H S+G FLTHCGWNST+ES+ + VPM+ +P DQ TN + I + W IG+
Sbjct: 1066SWCPQEKVLNHPSVGGFLTHCGWNSTIESICAGVPMLCWPFVADQPTNCRYICNEWDIGI 1245
Query: 409 RVDLHVNEDGIVEGEEIRRGLTVVMKNGEKGEELRRNAKKWKGLAKE 455
VD +V + E E + L V G+KG+++R+ + K A+E
Sbjct: 1246EVDTNVKRE---EVENLIHELMV----GDKGKKMRQRTMELKKKAEE 1365
>CB827201
Length = 374
Score = 143 bits (360), Expect = 7e-35
Identities = 70/113 (61%), Positives = 86/113 (75%), Gaps = 1/113 (0%)
Frame = +2
Query: 9 MVSHRFLIVSYPGQGQINPTLQFSKRLIAMGA-HVTIVTTLHMLRRMTTKTTIPGLSYTA 67
M HRFL++ YP QG INPTLQF+KRLI+ GA HVT+ TTLH RR++ K T+P L+
Sbjct: 32 MPHHRFLLIIYPAQGHINPTLQFAKRLISSGAEHVTLCTTLHTYRRISNKPTLPNLTLLP 211
Query: 68 FSDGFDDGFIVDVATHFNLYTPELKRRGSEFITNLILFAENEGHPFTCLIHTL 120
FSDGFDDGF + T ++LY ELKRRG+EF+T+LIL A EG PFTCLI+TL
Sbjct: 212 FSDGFDDGFKLTPDTDYSLYATELKRRGTEFVTDLILSAAQEGKPFTCLIYTL 370
>TC16757 weakly similar to UP|Q9SBQ2 (Q9SBQ2) Anthocyanin
5-O-glucosyltransferase, partial (19%)
Length = 638
Score = 125 bits (315), Expect = 1e-29
Identities = 62/86 (72%), Positives = 73/86 (84%)
Frame = +2
Query: 374 TMESLVSRVPMVAFPQWTDQNTNAKLIEDVWKIGVRVDLHVNEDGIVEGEEIRRGLTVVM 433
TMESL S VPM AFP+++DQ TNAK++EDVWKIGVR+D VNEDG+VEG EIR+ L VM
Sbjct: 2 TMESLXSGVPMXAFPRFSDQMTNAKMVEDVWKIGVRMDHSVNEDGVVEGGEIRKCLDAVM 181
Query: 434 KNGEKGEELRRNAKKWKGLAKEAGKE 459
NGEK EE+RRNA+K+KGLA EAGKE
Sbjct: 182 GNGEKAEEVRRNAEKFKGLAMEAGKE 259
>TC9836 weakly similar to UP|Q9LXV0 (Q9LXV0) Glucosyltransferase-like
protein, partial (55%)
Length = 1286
Score = 117 bits (293), Expect = 4e-27
Identities = 66/190 (34%), Positives = 110/190 (57%), Gaps = 4/190 (2%)
Frame = +1
Query: 274 EWLDSKPPSSVVYVSFGSYIVLSKTQMDEIARALLDCRHPFLWVIREKEKFQDGEKEKEE 333
+WLD+KP +SV++VSFGS +S +QM ++A AL F+WV+R F + + E
Sbjct: 385 KWLDTKPSNSVLFVSFGSMNTISASQMMQLATALDRSGRNFIWVVRPPIGFDINSEFRAE 564
Query: 334 ELGNIEELEEVGK----IAKWCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQ 389
E L V K + W QVE+LSH ++ FL+HCGWNS +ESL VP++ +P
Sbjct: 565 EWLPEGFLGRVAKKGLVVHDWAPQVEILSHGAVSAFLSHCGWNSVLESLSHGVPILGWPM 744
Query: 390 WTDQNTNAKLIEDVWKIGVRVDLHVNEDGIVEGEEIRRGLTVVMKNGEKGEELRRNAKKW 449
+Q N K++E+ ++GV V++ + V E++ + +VM E G ++R+NA
Sbjct: 745 AAEQFFNCKMLEE--ELGVCVEVARGKRCEVRHEDLVEKIELVMNEAESGVKIRKNAGNI 918
Query: 450 KGLAKEAGKE 459
+ + ++A ++
Sbjct: 919 REMIRDAVRD 948
>TC9827 weakly similar to UP|Q8S9A2 (Q8S9A2) Glucosyltransferase-7
(Fragment), partial (42%)
Length = 611
Score = 115 bits (288), Expect = 2e-26
Identities = 58/117 (49%), Positives = 76/117 (64%)
Frame = +2
Query: 340 ELEEVGKIAKWCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQNTNAKL 399
E E G I WC Q+ VL H+++GCFLTHCGWNST+ES+ VP++A P WTD TN KL
Sbjct: 11 ETTEKGLIVTWCPQLLVLKHQALGCFLTHCGWNSTLESVSLGVPVIAMPLWTDPVTNGKL 190
Query: 400 IEDVWKIGVRVDLHVNEDGIVEGEEIRRGLTVVMKNGEKGEELRRNAKKWKGLAKEA 456
I DVWK GVR +E IV E I+ + +++ EK E++ NA KW LAK++
Sbjct: 191 IVDVWKTGVRA--VADEKEIVRRETIKNCIKEIIET-EKINEIKSNAIKWMNLAKDS 352
>CB829602
Length = 542
Score = 110 bits (276), Expect = 4e-25
Identities = 60/152 (39%), Positives = 89/152 (58%), Gaps = 1/152 (0%)
Frame = +1
Query: 273 IEWLDSKPPSSVVYVSFGSYIVLSKTQMDEIARALLDCRHPFLWVIREKEKFQDGEKEKE 332
++WLDS+ PSSV+YV+FGS I ++ Q+ E+A + + + F+WVIR E E
Sbjct: 97 LKWLDSQEPSSVLYVNFGSVINMTPQQLVELAWGIANSKKKFIWVIRPDLV----EGEAS 264
Query: 333 EELGNI-EELEEVGKIAKWCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWT 391
L I E ++ G + WC Q +VL H ++G FLTHCGWNST+ES+ S VP++ P +
Sbjct: 265 IVLPEIVAETKDRGIMLSWCPQEQVLKHSALGGFLTHCGWNSTIESISSGVPLICSPFFN 444
Query: 392 DQNTNAKLIEDVWKIGVRVDLHVNEDGIVEGE 423
DQ N++ I W G + +N D + E
Sbjct: 445 DQFVNSRYICSEWDFG----MEMNSDDVKRDE 528
>TC14237 weakly similar to UP|AAR06913 (AAR06913) UDP-glycosyltransferase
85A8, partial (27%)
Length = 999
Score = 110 bits (275), Expect = 5e-25
Identities = 56/144 (38%), Positives = 85/144 (58%)
Frame = +2
Query: 313 PFLWVIREKEKFQDGEKEKEEELGNIEELEEVGKIAKWCSQVEVLSHRSMGCFLTHCGWN 372
PFLW++R + +E ++E++ G IA WC Q EVLSH S+G FLTHCGWN
Sbjct: 215 PFLWILRPDVVMGEDSINVPQEF--LDEIKGRGYIASWCFQEEVLSHPSIGAFLTHCGWN 388
Query: 373 STMESLVSRVPMVAFPQWTDQNTNAKLIEDVWKIGVRVDLHVNEDGIVEGEEIRRGLTVV 432
ST+E + + +P++ +P +++Q+TN++ W+IG+ V+ V D I L
Sbjct: 389 STIEGISAGLPLICWPFFSEQHTNSRYACTTWEIGMEVNHDVKRDEITT-------LVNE 547
Query: 433 MKNGEKGEELRRNAKKWKGLAKEA 456
M GEKG+E+R+ +WK A EA
Sbjct: 548 MMKGEKGKEMRKKGLEWKKKAIEA 619
>TC18079 weakly similar to UP|UFO5_MANES (Q40287) Flavonol
3-O-glucosyltransferase 5 (UDP-glucose flavonoid
3-O-glucosyltransferase 5) , partial (33%)
Length = 889
Score = 106 bits (264), Expect = 9e-24
Identities = 67/197 (34%), Positives = 109/197 (55%), Gaps = 10/197 (5%)
Frame = +3
Query: 270 NDYIEWLDSKPPSSVVYVSFGSYIVLSKTQMDEIARALLDCRHPFLWVIREKEK------ 323
++ + WLD +P SV+YVSFGS + + QM EIA L +H F+WV+R +
Sbjct: 54 DEILRWLDKQPAGSVIYVSFGSGGTMPQGQMTEIALGLELSQHRFVWVVRPPNEADASAT 233
Query: 324 -FQDGEKEKEEEL--GNIEELEEVGKIAK-WCSQVEVLSHRSMGCFLTHCGWNSTMESLV 379
F G + L G + E G + W Q E+L H + G F+T CGWNS +ES++
Sbjct: 234 FFGFGNEMALNYLPEGFMNRTREFGVVVPMWAPQAEILGHPATGGFVTPCGWNSVLESVL 413
Query: 380 SRVPMVAFPQWTDQNTNAKLIEDVWKIGVRVDLHVNEDGIVEGEEIRRGLTVVMKNGEKG 439
+ VPMVA+P +++Q NA ++ + ++GV V + V E G+V +++ + VM + E+G
Sbjct: 414 NGVPMVAWPLYSEQKMNAYMLSE--ELGVAVRVKVAEGGVVCRDQVVDMVRRVMVS-EEG 584
Query: 440 EELRRNAKKWKGLAKEA 456
++ ++ K EA
Sbjct: 585 MGMKDRVRELKLSGDEA 635
>TC19068 weakly similar to UP|AAR06913 (AAR06913) UDP-glycosyltransferase
85A8, partial (26%)
Length = 565
Score = 96.7 bits (239), Expect = 7e-21
Identities = 50/118 (42%), Positives = 75/118 (63%)
Frame = +1
Query: 338 IEELEEVGKIAKWCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQNTNA 397
I E+ G IA WCSQ VL+H S+G FLTHCGWNST+ES+ + VPM+ +P + DQ TN
Sbjct: 31 INEISSRGLIASWCSQEHVLNHPSIGGFLTHCGWNSTIESISAGVPMLCWPFFADQLTNC 210
Query: 398 KLIEDVWKIGVRVDLHVNEDGIVEGEEIRRGLTVVMKNGEKGEELRRNAKKWKGLAKE 455
+ I + W IG+++D + EE+ + + +M GEKG+++R+ + K A+E
Sbjct: 211 RSICNEWDIGMQIDTNGKR------EEVEKLINELMV-GEKGKKMRQKTMELKKRAEE 363
>TC9485 similar to UP|Q8S999 (Q8S999) Glucosyltransferase-10, partial (75%)
Length = 1195
Score = 96.3 bits (238), Expect = 1e-20
Identities = 95/379 (25%), Positives = 162/379 (42%), Gaps = 17/379 (4%)
Frame = +1
Query: 7 TAMVSHRFLIVSYPGQGQINPTLQFSKRLIAMGA-HVTIVTTLHMLRRMTTK------TT 59
TA + + YP QG INP L+ +K L G HVT V T + +R+
Sbjct: 73 TAAKKPHVVCIPYPAQGHINPMLKLAKLLHFKGGFHVTFVNTEYNHKRLLKSRGPDSLNG 252
Query: 60 IPGLSYTAFSDGFDDGFIVDVATHFNLYTPELKRRGSEFITNLILF---AENEGHPFTCL 116
+P + DG + VDV ++ L+ ++ P TC+
Sbjct: 253 LPSFRFETIPDGLPETD-VDVTQDIPSLCISTRKTCLPHFKKLLSKLNDVSSDVPPVTCI 429
Query: 117 IHTLFVPWAPQVARELNLITAKLWIEPTTVMS--ILYNYFLGGDYIAKKNTVESS----- 169
+ + + A ELN+ W + Y + I K++ + +
Sbjct: 430 VSDGCMSFTLDAAIELNIPEVLFWTTSACGFMGYVQYRELIEKGIIPLKDSSDITNGYLE 609
Query: 170 CSIEFSGLPFSLSPHDMPSYLISKGSYVLPLFKEDFKELRG*ANTTILVNTFEALEPEAP 229
+IE+ ++ D+PS+L + L + R + I++NTF+ALE +
Sbjct: 610 TTIEWLPGMKNIRLKDLPSFLRTTDPNDKMLDFLTGECQRALKASAIILNTFDALEHDVL 789
Query: 230 RAVDKLNMIPIGLLIPSAFLDRKDFAHDTSFGGDIIQAPSNDYIEWLDSKPPSSVVYVSF 289
A + + P+ + P L + + + G + ++ ++WLD+K P+SVVYV+F
Sbjct: 790 EAFSSI-LPPVYSIGPLHLLIKDVTDKNLNSLGSNLWKEDSECLKWLDTKEPNSVVYVNF 966
Query: 290 GSYIVLSKTQMDEIARALLDCRHPFLWVIREKEKFQDGEKEKEEELGNIEELEEVGKIAK 349
GS V++ QM E A L + FLWVIR + EE + + G+++
Sbjct: 967 GSITVMTSEQMVEFAWGLANSNKTFLWVIR-PDLVAGKHAVLPEEF--VAATNDRGRLSS 1137
Query: 350 WCSQVEVLSHRSMGCFLTH 368
W Q +VL+H ++G FLTH
Sbjct: 1138WTPQEDVLTHPAIGGFLTH 1194
>BI421006
Length = 519
Score = 91.7 bits (226), Expect = 2e-19
Identities = 52/144 (36%), Positives = 84/144 (58%), Gaps = 5/144 (3%)
Frame = +2
Query: 282 SSVVYVSFGSYIVLSKTQMDEIARALLDCRHPFLWVIREKEKFQ--DGEKEKEEEL--GN 337
S ++ F ++I + Q+++IA L + F+WV+R+ +K DG+K KE L G
Sbjct: 89 SICIFWFFSTFI---EEQIEQIANGLEQSKQKFIWVLRDADKGDIFDGDKVKERGLPKGF 259
Query: 338 IEELEEVGKIAK-WCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQNTN 396
+ +E +G + + W Q+E+LSH S G F++HCGWNS +ES+ VP+ A+P +DQ N
Sbjct: 260 EKRVEGMGLVVRDWAPQLEILSHPSTGGFMSHCGWNSCIESMSMGVPIAAWPMHSDQPRN 439
Query: 397 AKLIEDVWKIGVRVDLHVNEDGIV 420
LI +V K+ + V D +V
Sbjct: 440 TVLITEVLKVALVVQDXAQRDELV 511
Score = 29.3 bits (64), Expect = 1.5
Identities = 12/23 (52%), Positives = 16/23 (69%)
Frame = +3
Query: 270 NDYIEWLDSKPPSSVVYVSFGSY 292
N +EWLD + SV+YVSFG +
Sbjct: 45 NFVMEWLDRQEQKSVLYVSFGFF 113
>TC10667 similar to UP|Q9M051 (Q9M051) Glucuronosyl transferase-like
protein, partial (20%)
Length = 824
Score = 88.6 bits (218), Expect = 2e-18
Identities = 46/108 (42%), Positives = 66/108 (60%)
Frame = +1
Query: 343 EVGKIAKWCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQNTNAKLIED 402
E G KW Q +VL H ++G F THCGWNST+ES+ VPM+ P + DQ NAK + D
Sbjct: 1 EFGTSVKWAPQEQVLKHPAIGAFWTHCGWNSTLESVCEGVPMICSPCFGDQKVNAKYVSD 180
Query: 403 VWKIGVRVDLHVNEDGIVEGEEIRRGLTVVMKNGEKGEELRRNAKKWK 450
VWK+GV++ N+ G EI + ++++M G++ E+R N K K
Sbjct: 181 VWKVGVQLQ---NKPG---RGEIEKTISLLML-GDEAYEIRGNILKLK 303
>TC16143 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltransferase,
partial (30%)
Length = 604
Score = 87.4 bits (215), Expect = 5e-18
Identities = 45/120 (37%), Positives = 72/120 (59%), Gaps = 7/120 (5%)
Frame = +1
Query: 347 IAKWCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQNTNAKLIEDVWKI 406
I W QV +L HR++G F+THCGWNST+E++ + VPM+ +P +Q N KLI +V I
Sbjct: 19 IRGWVPQVVILGHRAVGAFVTHCGWNSTVEAVSAGVPMITWPVVGEQFYNEKLITEVRGI 198
Query: 407 GVRVDLHV-------NEDGIVEGEEIRRGLTVVMKNGEKGEELRRNAKKWKGLAKEAGKE 459
GV V + +V E I + + +M G++GE++RR A+++ A++A +E
Sbjct: 199 GVEVGAEECCLMGFDKREKLVSRESIEKAVRRLMDGGDEGEQIRRRAQEYGEKARQAVEE 378
>TC10408 weakly similar to UP|Q9FUJ6 (Q9FUJ6) UDP-glucosyltransferase HRA25,
partial (19%)
Length = 571
Score = 86.3 bits (212), Expect = 1e-17
Identities = 44/106 (41%), Positives = 67/106 (62%)
Frame = +1
Query: 345 GKIAKWCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQNTNAKLIEDVW 404
GK+ KW Q ++L+H ++ CF++HCGWNST+E + + VP + +P TDQ N I DVW
Sbjct: 16 GKVVKWGPQKKILNHPAIACFISHCGWNSTIEGVHASVPFLCWPFSTDQFLNKSFICDVW 195
Query: 405 KIGVRVDLHVNEDGIVEGEEIRRGLTVVMKNGEKGEELRRNAKKWK 450
KIG+ +D +E+GI+ EIR + V+ N E+L+ + K K
Sbjct: 196 KIGLGLD--KDENGIIPKGEIRTKVEQVIVN----EDLKARSLKLK 315
>TC8914 similar to UP|Q9ZWS2 (Q9ZWS2) Flavonoid 3-O-galactosyl transferase,
partial (32%)
Length = 627
Score = 86.3 bits (212), Expect = 1e-17
Identities = 43/121 (35%), Positives = 70/121 (57%)
Frame = +2
Query: 336 GNIEELEEVGKIAKWCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQNT 395
G IE + GKI W Q ++L H S+G F+THCG NS ES+ + VPM+ P + D
Sbjct: 38 GFIERTKNSGKIVPWAPQTQILGHESVGVFVTHCGCNSVFESISNGVPMICRPFFGDHGM 217
Query: 396 NAKLIEDVWKIGVRVDLHVNEDGIVEGEEIRRGLTVVMKNGEKGEELRRNAKKWKGLAKE 455
++IED+W+IGV++ E G+ + + + L +++ E+G+ +R A+K K +
Sbjct: 218 AGRIIEDIWEIGVKI-----EGGVFTQDGLVKSLNLILVQ-EEGKIMREKAQKVKKTVLD 379
Query: 456 A 456
A
Sbjct: 380 A 382
>TC9184 weakly similar to UP|HQGT_RAUSE (Q9AR73) Hydroquinone
glucosyltransferase (Arbutin synthase) , partial (26%)
Length = 643
Score = 83.6 bits (205), Expect = 7e-17
Identities = 47/107 (43%), Positives = 69/107 (63%)
Frame = +3
Query: 353 QVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQNTNAKLIEDVWKIGVRVDL 412
Q +VLSH S+G FL+HCGWNS +ES+ + VP+VA+P + +Q NA L+ + K+ +R
Sbjct: 9 QAKVLSHASIGGFLSHCGWNSVLESVANGVPLVAWPLYAEQRMNAVLVTEDVKVALRP-- 182
Query: 413 HVNEDGIVEGEEIRRGLTVVMKNGEKGEELRRNAKKWKGLAKEAGKE 459
V E+G+VE EI R + +M+ GE+G++LR K K A KE
Sbjct: 183 KVGENGLVELVEIARVVRTLMQ-GEEGKKLRCKMKDLKEAAATTLKE 320
>TC17165 similar to UP|Q9SMG6 (Q9SMG6) Betanidin-5-O-glucosyltransferase,
partial (24%)
Length = 546
Score = 81.6 bits (200), Expect = 2e-16
Identities = 46/118 (38%), Positives = 68/118 (56%), Gaps = 5/118 (4%)
Frame = +1
Query: 347 IAKWCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQNTNAKLIEDVWKI 406
I W QV +L H ++G F+THCGWNST+E + + VPMV +P +Q N KL+ +V KI
Sbjct: 4 IRGWAPQVLILEHEAIGAFVTHCGWNSTLEGVAAGVPMVTWPIAAEQFYNEKLVTEVLKI 183
Query: 407 GVRVDLH-----VNEDGIVEGEEIRRGLTVVMKNGEKGEELRRNAKKWKGLAKEAGKE 459
GV V V E V+ + + +G+ +M+ GE+ EE+R K A+ A +E
Sbjct: 184 GVPVGAKKWGRMVLEGDGVKSDAVEKGVKRIME-GEEAEEMRNRVKVLSQQAQWAVEE 354
>BP039584
Length = 545
Score = 80.9 bits (198), Expect = 4e-16
Identities = 39/112 (34%), Positives = 68/112 (59%), Gaps = 6/112 (5%)
Frame = -3
Query: 350 WCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQNTNAKLIEDVWKIGVR 409
W Q+ +L H ++G +THCGWN+T+ES+++ +PM P + +Q N KL+ DV +GV
Sbjct: 537 WAPQLLILEHPAIGGVVTHCGWNTTLESVIAGLPMATMPLFAEQFYNEKLLVDVLGVGVS 358
Query: 410 VDL------HVNEDGIVEGEEIRRGLTVVMKNGEKGEELRRNAKKWKGLAKE 455
+ + +V D IV+ E I + ++++M GE+ E+RR ++ AK+
Sbjct: 357 IGMKKWRNWNVVGDEIVKRENIVKAISLLMGGGEEALEMRRRVRELSDAAKK 202
>CN825072
Length = 712
Score = 80.9 bits (198), Expect = 4e-16
Identities = 52/184 (28%), Positives = 96/184 (51%), Gaps = 16/184 (8%)
Frame = -3
Query: 290 GSYIVLSKTQMDEIARALLDCRHPFLWVIRE---KEKFQDGEKEKEEEL------GNIEE 340
GS + Q+ EIARA+ + F+W +R+ K ++L G ++
Sbjct: 710 GSRGSFDRAQITEIARAVENSGVRFVWSLRKPXLKGSMVGPSDYSVDDLASVLPEGFLDR 531
Query: 341 LEEVGKIAKWCSQVEVLSHRSMGCFLTHCGWNSTMESLVSRVPMVAFPQWTDQNTNAKLI 400
+G++ W Q VL+H + G F++HCGWNST+ES+ VP+ +P + +Q TNA ++
Sbjct: 530 TTGIGRVIGWAPQTRVLTHPATGGFVSHCGWNSTLESIYFGVPIATWPLYAEQQTNAFVL 351
Query: 401 EDVWKIGV------RVDLHVNEDGIVEGEEIRRGL-TVVMKNGEKGEELRRNAKKWKGLA 453
KI V RV+++ + ++ ++I G+ +V+ K+GE + ++ ++K +
Sbjct: 350 VRELKIAVEISLDYRVEINGGPNYLLTADKIEGGIRSVLDKDGEVRKRVKEMSEKSRKTL 171
Query: 454 KEAG 457
E G
Sbjct: 170 LEGG 159
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.323 0.139 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,222,405
Number of Sequences: 28460
Number of extensions: 110511
Number of successful extensions: 648
Number of sequences better than 10.0: 96
Number of HSP's better than 10.0 without gapping: 626
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 629
length of query: 475
length of database: 4,897,600
effective HSP length: 94
effective length of query: 381
effective length of database: 2,222,360
effective search space: 846719160
effective search space used: 846719160
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0108.9


