

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0108.8
(935 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
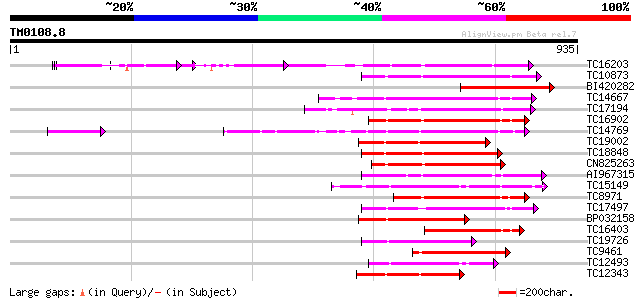
Score E
Sequences producing significant alignments: (bits) Value
TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodula... 274 5e-74
TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partia... 231 4e-61
BI420282 213 1e-55
TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, com... 212 2e-55
TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete 211 6e-55
TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-... 209 2e-54
TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete 204 4e-53
TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866)... 202 2e-52
TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, pa... 200 1e-51
CN825263 196 1e-50
AI967315 177 7e-45
TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like pro... 171 6e-43
TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P... 164 6e-41
TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2,... 157 6e-39
BP032158 146 2e-35
TC16403 homologue to UP|Q9LKY4 (Q9LKY4) Pti1 kinase-like protein... 145 3e-35
TC19726 similar to UP|Q9XIS3 (Q9XIS3) Lectin-like protein kinase... 144 6e-35
TC9461 similar to UP|O49840 (O49840) Protein kinase (AT2G02800/T... 143 1e-34
TC12493 similar to UP|Q9LP77 (Q9LP77) T1N15.9, partial (35%) 140 7e-34
TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine prote... 140 7e-34
>TC16203 UP|Q8GRU6 (Q8GRU6) LRR receptor-like kinase (Hypernodulation aberrant
root formation protein), complete
Length = 3308
Score = 274 bits (700), Expect = 5e-74
Identities = 239/833 (28%), Positives = 378/833 (44%), Gaps = 46/833 (5%)
Frame = +2
Query: 77 ISLASRSLTGTLPSDLNSLSQLTSLSL-QNNAISGPIP-SLANLSALKTAFLGRNNFTSV 134
+ L + SLTG +P L L L L L +NA G IP + ++ L+ + N T
Sbjct: 707 LGLNANSLTGRVPESLAKLKTLKELHLGYSNAYEGGIPPAFGSMENLRLLEMANCNLTGE 886
Query: 135 PSASFAGLTDLQTLSLSDNPNLSPWTLPTELTQSTNLITLELGTARLTGQLPESFF---- 190
S LT L +L + N NL+ T+P EL+ +L++L+L LTG++PESF
Sbjct: 887 IPPSLGNLTKLHSLFVQMN-NLTG-TIPPELSSMMSLMSLDLSINDLTGEIPESFSKLKN 1060
Query: 191 --------DKF-----------PGLQSVRLSYNNLTGALPNSLAASAIENLWLNNQDNGL 231
+KF P L+++++ NN + LP++L + L+ + N L
Sbjct: 1061 LTLMNFFQNKFRGSLPSFIGDLPNLETLQVWENNFSFVLPHNLGGNG-RFLYFDVTKNHL 1237
Query: 232 SGTIDV-LSNMTQLAQVWLHKNQFTGPIPD-LSQCSNLFDLQLRDNQLTGPVPNSLMGLT 289
+G I L +L + N F GPIP + +C +L +++ +N L GPVP + L
Sbjct: 1238 TGLIPPDLCKSGRLKTFIITDNFFRGPIPKGIGECRSLTKIRVANNFLDGPVPPGVFQLP 1417
Query: 290 SLQNVSLDNNELQGPFPAFGKGVKV---TLDGINSFCKDTPGPC-----------DARVM 335
S+ L NN L G P+ G + TL N F P DA
Sbjct: 1418 SVTITELSNNRLNGELPSVISGESLGTLTLSN-NLFTGKIPAAMKNLRALQSLSLDANEF 1594
Query: 336 VLLHIAGAFGYPIKFAGSWKGNDPCQGWSFVVCDSGRKIITVNLAKQGLQGTISPAFANL 395
+ G F P+ + GN+ G + V+L++ L G + NL
Sbjct: 1595 IGEIPGGVFEIPMLTKVNISGNN-LTGPIPTTITHRASLTAVDLSRNNLAGEVPKGMKNL 1771
Query: 396 TDLRSLYLNGNNLTGSIPESLTTLTQLETLEVSDNNLSGEVPKFPPKVKLLTAGNVLLGQ 455
DL L L+ N ++G +P+ + +T L TL++S NN +G VP T G L+
Sbjct: 1772 MDLSILNLSRNEISGPVPDEIRFMTSLTTLDLSSNNFTGTVP---------TGGQFLVFN 1924
Query: 456 TTGSGGGAGGKTTPSAGSTPEGSHGESGNGSSLTPGWIAGIVIIVLFFVAVVLFVSCKCY 515
+ G P S P + + T A ++ I L +++ V+
Sbjct: 1925 YDKTFAGNPNLCFPHRASCPSVLYDSLRKTRAKTARVRAIVIGIALATAVLLVAVTVHVV 2104
Query: 516 AKRRHGKFSRVANPVNGNGNVKLDVISVSNGYSGAPSELQSQSSGDHSELHVFDGGNSTM 575
KRR + ++ ++ E+ D
Sbjct: 2105 RKRRLHR-----------------------------AQAWKLTAFQRLEIKAED------ 2179
Query: 576 SILVLRQVTGNFSEDNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEIT 635
V E+NI+G+GG G+VY+G + +GT +A+KR+ G F+AEI
Sbjct: 2180 -------VVECLKEENIIGKGGAGIVYRGSMPNGTDVAIKRLVGQGSGRNDYG-FRAEIE 2335
Query: 636 VLSKVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVAL 695
L K+RHR+++ LLG+ N + LL+YEYMP G+L + L + G+ L W+ R +A+
Sbjct: 2336 TLGKIRHRNIMRLLGYVSNKDTNLLLYEYMPNGSLGEWLHGAKG-GH--LRWEMRYKIAV 2506
Query: 696 DVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPD-GKYSVETRLAG 754
+ ARG+ Y+H IHRD+K +NILL D A VADFGL K D G + +AG
Sbjct: 2507 EAARGLCYMHHDCSPLIIHRDVKSNNILLDADFEAHVADFGLAKFLYDPGASQSMSSIAG 2686
Query: 755 TFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVL--I 812
++GY+APEYA T +V K DVY+FGVVL+ELI GR+ + + + +V W + + +
Sbjct: 2687 SYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIIGRKPVGEF--GDGVDIVGWVNKTMSEL 2860
Query: 813 NKENIPKAIDQTLNP--DEETMESIYKVSELAGHCTAREPNQRPDMGHAVNVL 863
++ + + ++P + S+ + +A C RP M V++L
Sbjct: 2861 SQPSDTALVLAVVDPRLSGYPLTSVIHMFNIAMMCVKEMGPARPTMREVVHML 3019
Score = 102 bits (254), Expect = 3e-22
Identities = 88/296 (29%), Positives = 137/296 (45%), Gaps = 3/296 (1%)
Frame = +2
Query: 167 QSTNLITLELGTARLTGQLPESFFDKFPGLQSVRLSYNNLTGALPNSLAASAIENLWLNN 226
Q+ ++ L + L G LP L+++ +S NNLT LP+ LA+ + LN
Sbjct: 323 QNLRVVALNVTLVPLFGHLPPEI-GLLEKLENLTISMNNLTDQLPSDLASLTSLKV-LNI 496
Query: 227 QDNGLSGTI--DVLSNMTQLAQVWLHKNQFTGPIPD-LSQCSNLFDLQLRDNQLTGPVPN 283
N SG ++ MT+L + + N F+GP+P+ + + L L L N +G +P
Sbjct: 497 SHNLFSGQFPGNITVGMTELEALDAYDNSFSGPLPEEIVKLEKLKYLHLAGNYFSGTIPE 676
Query: 284 SLMGLTSLQNVSLDNNELQGPFPAFGKGVKVTLDGINSFCKDTPGPCDARVMVLLHIAGA 343
S SL+ + L+ N L G P S K + + LH+
Sbjct: 677 SYSEFQSLEFLGLNANSLTGRVP-------------ESLAK-------LKTLKELHL--- 787
Query: 344 FGYPIKFAGSWKGNDPCQGWSFVVCDSGRKIITVNLAKQGLQGTISPAFANLTDLRSLYL 403
GY + G G P G S + + +A L G I P+ NLT L SL++
Sbjct: 788 -GYSNAYEG---GIPPAFG-------SMENLRLLEMANCNLTGEIPPSLGNLTKLHSLFV 934
Query: 404 NGNNLTGSIPESLTTLTQLETLEVSDNNLSGEVPKFPPKVKLLTAGNVLLGQTTGS 459
NNLTG+IP L+++ L +L++S N+L+GE+P+ K+K LT N + GS
Sbjct: 935 QMNNLTGTIPPELSSMMSLMSLDLSINDLTGEIPESFSKLKNLTLMNFFQNKFRGS 1102
Score = 80.9 bits (198), Expect = 9e-16
Identities = 63/242 (26%), Positives = 101/242 (41%), Gaps = 2/242 (0%)
Frame = +2
Query: 71 SNRVTTISLASRSLTGTLPSDLNSLSQLTSLSLQNNAISGPIP-SLANLSALKTAFLGRN 129
S R+ T + G +P + LT + + NN + GP+P + L ++ L N
Sbjct: 1268 SGRLKTFIITDNFFRGPIPKGIGECRSLTKIRVANNFLDGPVPPGVFQLPSVTITELSNN 1447
Query: 130 NFTSVPSASFAGLTDLQTLSLSDNPNLSPWTLPTELTQSTNLITLELGTARLTGQLPESF 189
+ +G L TL+LS+N L +P + L +L L G++P
Sbjct: 1448 RLNGELPSVISG-ESLGTLTLSNN--LFTGKIPAAMKNLRALQSLSLDANEFIGEIPGGV 1618
Query: 190 FDKFPGLQSVRLSYNNLTGALPNSLAASAIENLWLNNQDNGLSGTIDVLSNMTQLAQVWL 249
F+ P L V +S NNLTG +P ++ A L V L
Sbjct: 1619 FE-IPMLTKVNISGNNLTGPIPTTITHRA------------------------SLTAVDL 1723
Query: 250 HKNQFTGPIPD-LSQCSNLFDLQLRDNQLTGPVPNSLMGLTSLQNVSLDNNELQGPFPAF 308
+N G +P + +L L L N+++GPVP+ + +TSL + L +N G P
Sbjct: 1724 SRNNLAGEVPKGMKNLMDLSILNLSRNEISGPVPDEIRFMTSLTTLDLSSNNFTGTVPTG 1903
Query: 309 GK 310
G+
Sbjct: 1904 GQ 1909
Score = 78.6 bits (192), Expect = 4e-15
Identities = 61/211 (28%), Positives = 97/211 (45%), Gaps = 2/211 (0%)
Frame = +2
Query: 74 VTTISLASRSLTGTLPSDLNSLSQLTSLSLQNNAISGPIPSLANLSALKTAFLGRNNFTS 133
+T I +A+ L G +P + L +T L NN ++G +PS+ + +L T L N FT
Sbjct: 1349 LTKIRVANNFLDGPVPPGVFQLPSVTITELSNNRLNGELPSVISGESLGTLTLSNNLFTG 1528
Query: 134 VPSASFAGLTDLQTLSLSDNPNLSPWTLPTELTQSTNLITLELGTARLTGQLPESFFDKF 193
A+ L LQ+LSL N G++P F+
Sbjct: 1529 KIPAAMKNLRALQSLSLDAN--------------------------EFIGEIPGGVFE-I 1627
Query: 194 PGLQSVRLSYNNLTGALPNSLAASAIENLWLNNQDNGLSGTIDV-LSNMTQLAQVWLHKN 252
P L V +S NNLTG +P ++ A ++ N L+G + + N+ L+ + L +N
Sbjct: 1628 PMLTKVNISGNNLTGPIPTTITHRASLTA-VDLSRNNLAGEVPKGMKNLMDLSILNLSRN 1804
Query: 253 QFTGPIPD-LSQCSNLFDLQLRDNQLTGPVP 282
+ +GP+PD + ++L L L N TG VP
Sbjct: 1805 EISGPVPDEIRFMTSLTTLDLSSNNFTGTVP 1897
>TC10873 similar to UP|Q9LDZ5 (Q9LDZ5) F2D10.13 (F5M15.3), partial (77%)
Length = 1478
Score = 231 bits (589), Expect = 4e-61
Identities = 125/298 (41%), Positives = 175/298 (57%), Gaps = 1/298 (0%)
Frame = +1
Query: 580 LRQVTGNFSEDNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVLSK 639
L T NF E N++G GGFG VYKG L G +AVK++ G +G EF E+ +LS
Sbjct: 430 LADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHD--GRQGFQEFVMEVLMLSL 603
Query: 640 VRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDVAR 699
+ H +LV L+G+C +G++RLLVYEYMP G+L HLFE PL W R+ VA+ AR
Sbjct: 604 LHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSH-DKEPLNWSTRMKVAVGAAR 780
Query: 700 GVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYS-VETRLAGTFGY 758
G+EYLH A I+RDLK +NILL ++ K++DFGL K P G + V TR+ GT+GY
Sbjct: 781 GLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGY 960
Query: 759 LAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINKENIP 818
APEYA +G++T K D+Y+FGVVL+EL+TGRRA+D S +LV+W R ++
Sbjct: 961 CAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRRRFG 1140
Query: 819 KAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHAVNVLVPMVEQWKPTSRH 876
+D L +++ + C +P RP + V L + Q P + +
Sbjct: 1141HMVDPLLQ-GRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYLASQGSPEAHY 1311
>BI420282
Length = 504
Score = 213 bits (541), Expect = 1e-55
Identities = 99/155 (63%), Positives = 128/155 (81%)
Frame = +1
Query: 744 GKYSVETRLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHL 803
GK S ETRLAGTFGYLAPEYA TGRVTTKVDV++FGV+LMEL+TGR+ALD++ P++ HL
Sbjct: 37 GKASFETRLAGTFGYLAPEYAVTGRVTTKVDVFSFGVILMELLTGRKALDETQPEDSMHL 216
Query: 804 VTWFRRVLINKENIPKAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHAVNVL 863
VTWFRR+ I+K++ KAID ++ +EET+ S++ V+ELAGHC AREP QRPDMGHAVNVL
Sbjct: 217 VTWFRRMYIDKDSFRKAIDPSIEINEETLASVHTVAELAGHCCAREPYQRPDMGHAVNVL 396
Query: 864 VPMVEQWKPTSRHEDDGHDSEPHMSLPQVLQRWQA 898
+VE WKP+ ++ +D + + MSLPQ L++WQA
Sbjct: 397 SSLVELWKPSDQNSEDMYGIDLDMSLPQALKKWQA 501
>TC14667 homologue to UP|Q84P43 (Q84P43) Protein kinase Pti1, complete
Length = 1591
Score = 212 bits (540), Expect = 2e-55
Identities = 128/368 (34%), Positives = 211/368 (56%), Gaps = 8/368 (2%)
Frame = +2
Query: 509 FVSCKCYAKRRH--GKFSRVANPVN-GNGNVKLDVISVSNGYSGAPSELQSQSSGDHSEL 565
++ C C + + + + +P N G+GN K + AP + ++Q + E+
Sbjct: 158 WLCCTCQVEEPYPSNENDHLKSPRNYGDGNSK-------GSKASAPVKHETQKAPPPIEV 316
Query: 566 HVFDGGNSTMSILVLRQVTGNFSEDNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNK 625
+S+ L++ T NF ++G G +G VY L DG +AVK+++ V+ +
Sbjct: 317 -------PALSLDELKEKTDNFGSKALIGEGSYGRVYYATLNDGNAVAVKKLD-VSSEPE 472
Query: 626 GLNEFQAEITVLSKVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWREL-GYTP 684
NEF +++++S++++ + V L G+C+ GN R+L YE+ G+L L + + G P
Sbjct: 473 TNNEFLTQVSMVSRLKNDNFVELHGYCVEGNLRVLAYEFATMGSLHDILHGRKGVQGAQP 652
Query: 685 ---LTWKQRVTVALDVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNA 741
L W QRV +A+D ARG+EYLH Q + IHRD++ SN+L+ +D +AK+ADF L A
Sbjct: 653 GPTLDWIQRVRIAVDAARGLEYLHEKVQPAIIHRDIRSSNVLIFEDYKAKIADFNLSNQA 832
Query: 742 PDGKYSVE-TRLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDER 800
PD + TR+ GTFGY APEYA TG++T K DVY+FGVVL+EL+TGR+ +D ++P +
Sbjct: 833 PDMAARLHSTRVLGTFGYHAPEYAMTGQLTQKSDVYSFGVVLLELLTGRKPVDHTMPRGQ 1012
Query: 801 SHLVTWFRRVLINKENIPKAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHAV 860
LVTW ++++ + + +D L E + + K++ +A C E RP+M V
Sbjct: 1013QSLVTW-ATPRLSEDKVKQCVDPKLK-GEYPPKGVAKLAAVAALCVQYEAEFRPNMSIVV 1186
Query: 861 NVLVPMVE 868
L P+++
Sbjct: 1187KALQPLLK 1210
>TC17194 UP|CAE02590 (CAE02590) Nod-factor receptor 1b, complete
Length = 2193
Score = 211 bits (536), Expect = 6e-55
Identities = 142/392 (36%), Positives = 211/392 (53%), Gaps = 11/392 (2%)
Frame = +1
Query: 486 SSLTPGWIAGIVIIVLFFVAVVLFVSCKCYAKRRHGKFSRVANPVNGNGNVKLDVISVSN 545
+ L G GI I F + ++ F Y K+ K +++ ++ +S +
Sbjct: 790 AGLASGAAVGISIAGTFVLLLLAFCMYVRYQKKEEEK-AKLPTDISM-------ALSTQD 945
Query: 546 GYSGAPSELQ-SQSSGDHSE-----LHVFDGGNSTMSILVLRQVTGNFSEDNILGRGGFG 599
G + + +E + S SSG + + + S L + T NFS DN +G+GGFG
Sbjct: 946 GNASSSAEYETSGSSGPGTASATGLTSIMVAKSMEFSYQELAKATNNFSLDNKIGQGGFG 1125
Query: 600 VVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVLSKVRHRHLVALLGHCINGNERL 659
VY EL+ G K A+K+M+ A EF E+ VL+ V H +LV L+G+C+ G+
Sbjct: 1126 AVYYAELR-GKKTAIKKMDVQAS-----TEFLCELKVLTHVHHLNLVRLIGYCVEGS-LF 1284
Query: 660 LVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDVARGVEYLHSLAQQSFIHRDLKP 719
LVYE++ G L Q+L G PL W RV +ALD ARG+EY+H +IHRD+K
Sbjct: 1285 LVYEHIDNGNLGQYL---HGSGKEPLPWSSRVQIALDAARGLEYIHEHTVPVYIHRDVKS 1455
Query: 720 SNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAGTFGYLAPEYAATGRVTTKVDVYAFG 779
+NIL+ ++R KVADFGL K G +++TRL GTFGY+ PEYA G ++ K+DVYAFG
Sbjct: 1456 ANILIDKNLRGKVADFGLTKLIEVGNSTLQTRLVGTFGYMPPEYAQYGDISPKIDVYAFG 1635
Query: 780 VVLMELITGRRAL--DDSLPDERSHLVTWFRRVLINKENIPKAIDQTLNP---DEETMES 834
VVL ELI+ + A+ L E LV F L NK + A+ + ++P + ++S
Sbjct: 1636 VVLFELISAKNAVLKTGELVAESKGLVALFEEAL-NKSDPCDALRKLVDPRLGENYPIDS 1812
Query: 835 IYKVSELAGHCTAREPNQRPDMGHAVNVLVPM 866
+ K+++L CT P RP M V L+ +
Sbjct: 1813 VLKIAQLGRACTRDNPLLRPSMRSLVVALMTL 1908
>TC16902 homologue to UP|Q8VZH5 (Q8VZH5) Receptor protein kinase-like
protein, partial (46%)
Length = 941
Score = 209 bits (532), Expect = 2e-54
Identities = 117/267 (43%), Positives = 167/267 (61%), Gaps = 1/267 (0%)
Frame = +3
Query: 592 ILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVLSKVRHRHLVALLGH 651
+LG GGFG VY GE+ GTK+A+KR ++ +G++EFQ EI +LSK+RHRHLV+L+G+
Sbjct: 12 LLGVGGFGKVYYGEVDGGTKVAIKRGNPLS--EQGVHEFQTEIEMLSKLRHRHLVSLIGY 185
Query: 652 CINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDVARGVEYLHSLAQQS 711
C E +LVY++M GTL +HL++ ++ PL WKQR+ + + ARG+ YLH+ A+ +
Sbjct: 186 CEENTEMILVYDHMAYGTLREHLYKTQK---PPLPWKQRLEICIGAARGLHYLHTGAKYT 356
Query: 712 FIHRDLKPSNILLGDDMRAKVADFGLVKNAPD-GKYSVETRLAGTFGYLAPEYAATGRVT 770
IHRD+K +NILL + AKV+DFGL K P V T + G+FGYL PEY ++T
Sbjct: 357 IIHRDVKTTNILLDEKWVAKVSDFGLSKTGPTLDNTHVSTVVKGSFGYLDPEYFRRQQLT 536
Query: 771 TKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINKENIPKAIDQTLNPDEE 830
K DVY+FGVVL E++ R AL+ SL E+ L W NK + + +D L +
Sbjct: 537 DKSDVYSFGVVLFEILCARPALNPSLAKEQVSLAEWASH-CYNKGILDQILDPYLK-GKI 710
Query: 831 TMESIYKVSELAGHCTAREPNQRPDMG 857
E K +E A C + + +RP MG
Sbjct: 711 APECFKKFAETAMKCVSDQGIERPSMG 791
>TC14769 UP|Q8LKX1 (Q8LKX1) Receptor-like kinase SYMRK, complete
Length = 3495
Score = 204 bits (520), Expect = 4e-53
Identities = 163/513 (31%), Positives = 253/513 (48%), Gaps = 9/513 (1%)
Frame = +3
Query: 353 SWKGNDPC--QGWSFVVCD--SGRKIIT-VNLAKQGLQGTISPAFANLTDLRSLYLNGNN 407
SW G DPC W + CD +G +IT ++L+ L+G I + A +T+L +L ++ N+
Sbjct: 1524 SWSG-DPCILLPWKGIACDGSNGSSVITKLDLSSSNLKGLIPSSIAEMTNLETLNISHNS 1700
Query: 408 LTGSIPESLTTLTQLETLEVSDNNLSGEVPKFPPKVKLLTAGNVLLGQTTGSGGGAGGKT 467
GS+P S + L ++++S N+L G++P+ VKL ++ G
Sbjct: 1701 FDGSVP-SFPLSSLLISVDLSYNDLMGKLPE--SIVKLPHLKSLYFGCNEHMSPEDPANM 1871
Query: 468 TPSAGSTPEG--SHGESGNGSSLTPGWIA-GIVIIVLFFVAVVLFVSCKCYAKRRHGKFS 524
S +T G ES G + G I G ++I L F VLFV ++
Sbjct: 1872 NSSLINTDYGRCKGKESRFGQVIVIGAITCGSLLITLAFG--VLFVC----------RYR 2015
Query: 525 RVANPVNGNGNVKLDVISVSNGYSGAPSELQSQSSGDHSELHVFDGGNSTMSILVLRQVT 584
+ P G K + +N PS+ + + ++ + T
Sbjct: 2016 QKLIPWEGFAGKKYPM--ETNIIFSLPSK---------DDFFIKSVSIQAFTLEYIEVAT 2162
Query: 585 GNFSEDNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVLSKVRHRH 644
+ ++G GGFG VY+G L DG ++AVK S + +G EF E+ +LS ++H +
Sbjct: 2163 ERYK--TLIGEGGFGSVYRGTLNDGQEVAVKVRSSTS--TQGTREFDNELNLLSAIQHEN 2330
Query: 645 LVALLGHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDVARGVEYL 704
LV LLG+C ++++LVY +M G+L L+ L W R+++AL ARG+ YL
Sbjct: 2331 LVPLLGYCNESDQQILVYPFMSNGSLQDRLYG-EPAKRKILDWPTRLSIALGAARGLAYL 2507
Query: 705 HSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAP-DGKYSVETRLAGTFGYLAPEY 763
H+ +S IHRD+K SNILL M AKVADFG K AP +G V + GT GYL PEY
Sbjct: 2508 HTFPGRSVIHRDIKSSNILLDHSMCAKVADFGFSKYAPQEGDSYVSLEVRGTAGYLDPEY 2687
Query: 764 AATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINKENIPKAIDQ 823
T +++ K DV++FGVVL+E+++GR L+ P LV W I + + +D
Sbjct: 2688 YKTQQLSEKSDVFSFGVVLLEIVSGREPLNIKRPRTEWSLVEW-ATPYIRGSKVDEIVDP 2864
Query: 824 TLNPDEETMESIYKVSELAGHCTAREPNQRPDM 856
+ E++++V E+A C RP M
Sbjct: 2865 GIKGGYHA-EAMWRVVEVALQCLEPFSTYRPSM 2960
Score = 55.8 bits (133), Expect = 3e-08
Identities = 31/99 (31%), Positives = 51/99 (51%), Gaps = 3/99 (3%)
Frame = +3
Query: 63 WDGIKCDSSNR---VTTISLASRSLTGTLPSDLNSLSQLTSLSLQNNAISGPIPSLANLS 119
W GI CD SN +T + L+S +L G +PS + ++ L +L++ +N+ G +PS S
Sbjct: 1557 WKGIACDGSNGSSVITKLDLSSSNLKGLIPSSIAEMTNLETLNISHNSFDGSVPSFPLSS 1736
Query: 120 ALKTAFLGRNNFTSVPSASFAGLTDLQTLSLSDNPNLSP 158
L + L N+ S L L++L N ++SP
Sbjct: 1737 LLISVDLSYNDLMGKLPESIVKLPHLKSLYFGCNEHMSP 1853
>TC19002 weakly similar to UP|Q9XJ10 (Q9XJ10) ESTs C22458(C62866), partial
(48%)
Length = 780
Score = 202 bits (515), Expect = 2e-52
Identities = 102/218 (46%), Positives = 142/218 (64%)
Frame = +1
Query: 576 SILVLRQVTGNFSEDNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEIT 635
S+ L T NF+ DN LG GGFG VY G+L DG++IAVKR++ NK EF E+
Sbjct: 31 SLKELHSATNNFNYDNKLGEGGFGSVYWGQLWDGSQIAVKRLK--VWSNKADMEFAVEVE 204
Query: 636 VLSKVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVAL 695
+L++VRH++L++L G+C G ERL+VY+YMP +L HL + L W +R+ +A+
Sbjct: 205 ILARVRHKNLLSLRGYCAEGQERLIVYDYMPNLSLLSHL-HGQHSSECLLDWNRRMNIAI 381
Query: 696 DVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAGT 755
A G+ YLH A IHRD+K SN+LL D +A+VADFG K PDG V TR+ GT
Sbjct: 382 GSAEGIVYLHHQATPHIIHRDIKASNVLLDSDFQARVADFGFAKLIPDGATHVTTRVKGT 561
Query: 756 FGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALD 793
GYLAPEYA G+ DV++FG++L+EL +G++ L+
Sbjct: 562 LGYLAPEYAMLGKANECCDVFSFGILLLELASGKKPLE 675
>TC18848 weakly similar to UP|Q9SSQ6 (Q9SSQ6) F6D8.24 protein, partial (64%)
Length = 969
Score = 200 bits (508), Expect = 1e-51
Identities = 104/233 (44%), Positives = 146/233 (62%)
Frame = +2
Query: 580 LRQVTGNFSEDNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVLSK 639
L TG FS+DN LG GGFG VY G DG +IAVK+++ AM +K EF E+ VL +
Sbjct: 281 LHAATGGFSDDNKLGEGGFGSVYWGRTSDGLQIAVKKLK--AMNSKAEMEFAVEVEVLGR 454
Query: 640 VRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDVAR 699
VRH++L+ L G+C+ ++RL+VY+YMP +L HL + L W++R+ +A+ A
Sbjct: 455 VRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQFAVE-VQLNWQKRMKIAIGSAE 631
Query: 700 GVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAGTFGYL 759
G+ YLH IHRD+K SN+LL D VADFG K P+G + TR+ GT GYL
Sbjct: 632 GILYLHHEVTPHIIHRDIKASNVLLNSDFEPLVADFGFAKLIPEGVSHMTTRVKGTLGYL 811
Query: 760 APEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLI 812
APEYA G+V+ DVY+FG++L+EL+TGR+ + + LP +T + LI
Sbjct: 812 APEYAMWGKVSESCDVYSFGILLLELVTGRKPI-EKLPGGVKRTITEWAEPLI 967
>CN825263
Length = 663
Score = 196 bits (499), Expect = 1e-50
Identities = 107/223 (47%), Positives = 143/223 (63%), Gaps = 2/223 (0%)
Frame = +1
Query: 597 GFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVLSKVRHRHLVALLGHCINGN 656
GFG+VYKG L DG +AVK ++ +G EF AE+ +LS++ HR+LV L+G CI
Sbjct: 1 GFGLVYKGILNDGRDVAVKILKRD--DQRGGREFLAEVEMLSRLHHRNLVKLIGICIEKQ 174
Query: 657 ERLLVYEYMPQGTLTQHLFEW-RELGYTPLTWKQRVTVALDVARGVEYLHSLAQQSFIHR 715
R L+YE +P G++ HL +E G PL W R+ +AL ARG+ YLH + IHR
Sbjct: 175 TRCLIYELVPNGSVESHLHGADKETG--PLDWNARMKIALGAARGLAYLHEDSNPCVIHR 348
Query: 716 DLKPSNILLGDDMRAKVADFGLVKNAPD-GKYSVETRLAGTFGYLAPEYAATGRVTTKVD 774
D K SNILL D KV+DFGL + A D G + T + GTFGYLAPEYA TG + K D
Sbjct: 349 DFKSSNILLECDFTPKVSDFGLARTALDEGNKHISTHVMGTFGYLAPEYAMTGHLLVKSD 528
Query: 775 VYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINKENI 817
VY++GVVL+EL+TG + +D S P + +LVTW R +L +KE +
Sbjct: 529 VYSYGVVLLELLTGTKPVDLSQPPGQENLVTWARPILTSKEGL 657
>AI967315
Length = 1308
Score = 177 bits (449), Expect = 7e-45
Identities = 112/309 (36%), Positives = 161/309 (51%), Gaps = 4/309 (1%)
Frame = +1
Query: 580 LRQVTGNFSEDNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVLSK 639
L T FS +N++G+GG+ VYKG L+ G +IAVKR+ + EF EI +
Sbjct: 127 LFHATNGFSSENMVGKGGYAEVYKGRLESGDEIAVKRLTRTCRDERKEKEFLTEIGTIGH 306
Query: 640 VRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDVAR 699
V H +++ LLG CI+ N LV+E G++ + + + PL WK R + L AR
Sbjct: 307 VCHSNVMPLLGCCID-NGLYLVFELSTVGSVASLIHDEK---MAPLDWKTRYKIVLGTAR 474
Query: 700 GVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAP-DGKYSVETRLAGTFGY 758
G+ YLH Q+ IHRD+K SNILL +D +++DFGL K P + + GTFG+
Sbjct: 475 GLHYLHKGCQRRIIHRDIKASNILLTEDFEPQISDFGLAKWLPSQWTHHSIAPIEGTFGH 654
Query: 759 LAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINKENIP 818
LAPEY G V K DV+AFGV L+E+I+GR+ +D S L TW + +L +K I
Sbjct: 655 LAPEYYMHGVVDEKTDVFAFGVFLLEVISGRKPVDGS----HQSLHTWAKPIL-SKWEIE 819
Query: 819 KAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHAVNVLVP---MVEQWKPTSR 875
K +D L + + +V+ A C RP M + V+ E+WK
Sbjct: 820 KLVDPRLEGCYDVTQ-FNRVAFAASLCIRASSTWRPTMSEVLEVMEEGEMDKERWKMPKE 996
Query: 876 HEDDGHDSE 884
E+D E
Sbjct: 997 EEEDQQGEE 1023
>TC15149 similar to UP|Q9LVI6 (Q9LVI6) Probable receptor-like protein kinase
protein (AT3g17840/MEB5_6), partial (45%)
Length = 1489
Score = 171 bits (432), Expect = 6e-43
Identities = 124/359 (34%), Positives = 187/359 (51%), Gaps = 3/359 (0%)
Frame = +3
Query: 531 NGNGNVKLDVISVSNGYSGAPSELQS-QSSGDHSELHVFDGGNSTMSIL--VLRQVTGNF 587
NGNGN NGYS A + + +++G ++ VF G ++ L +LR
Sbjct: 141 NGNGN--------GNGYSAAAATINKVEANGVGAKKLVFFGNSARGFDLEDLLR------ 278
Query: 588 SEDNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVLSKVRHRHLVA 647
+ +LG+G FG YK L+ G +AVKR++ V + K EF+ +I + + H+ LV
Sbjct: 279 ASAEVLGKGTFGTAYKAVLETGLVVAVKRLKDVTISEK---EFKDKIETVGAMDHQSLVP 449
Query: 648 LLGHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDVARGVEYLHSL 707
L + + +E+LLVY+YMP G+L+ L + G TPL W+ R +AL ARG+EYLHS
Sbjct: 450 LRAYYFSRDEKLLVYDYMPMGSLSALLHGNKGAGRTPLNWEIRSGIALGAARGIEYLHSQ 629
Query: 708 AQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAGTFGYLAPEYAATG 767
+ H ++K SNILL AKV+DFGL G S R+A GY APE
Sbjct: 630 G-PNVSHGNIKASNILLTKSYEAKVSDFGLAHLV--GPSSTPNRVA---GYRAPEVTDPR 791
Query: 768 RVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINKENIPKAIDQTLNP 827
+V+ K DVY+FGV+L+EL+TG+ L +E L W + L+ +E + D L
Sbjct: 792 KVSQKADVYSFGVLLLELLTGKAPTHALLNEEGVDLPRWVQS-LVREEWTSEVFDLELLR 968
Query: 828 DEETMESIYKVSELAGHCTAREPNQRPDMGHAVNVLVPMVEQWKPTSRHEDDGHDSEPH 886
+ E + ++ +LA C A P++RP + +E+ ++ ED D PH
Sbjct: 969 YQNVEEEMVQLLQLAVDCAAPYPDRRPSIAEVTR----SIEELCRSNLKED--QDQIPH 1127
>TC8971 similar to GB|AAM16251.1|20334780|AY093990 At2g11520/F14P14.15
{Arabidopsis thaliana;}, partial (37%)
Length = 1079
Score = 164 bits (415), Expect = 6e-41
Identities = 95/227 (41%), Positives = 141/227 (61%), Gaps = 3/227 (1%)
Frame = +3
Query: 633 EITVLSKVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVT 692
E+ +L+K+ HR+LV LLG GNER+L+ EY+P GTL +HL R L + QR+
Sbjct: 6 EVELLAKIDHRNLVKLLGFIDKGNERILITEYVPNGTLREHLDGLRG---KILDFNQRLE 176
Query: 693 VALDVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAP--DGKYSVET 750
+A+DVA G+ YLH A++ IHRD+K SNILL + MRAKVADFG + P + + T
Sbjct: 177 IAIDVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFARLGPVNGDQTHIST 356
Query: 751 RLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALD-DSLPDERSHLVTWFRR 809
++ GT GYL PEY T ++T K DVY+FG++L+E++TGRR ++ DER L FR+
Sbjct: 357 KVKGTVGYLDPEYMKTHQLTPKSDVYSFGILLLEILTGRRPVELKKAADERVTLRWAFRK 536
Query: 810 VLINKENIPKAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDM 856
N+ ++ + +D L + + + K+ +L+ C A RP+M
Sbjct: 537 --YNEGSVVELMD-PLMEEAVNADVLMKMLDLSFQCAAPIRTDRPNM 668
>TC17497 UP|CAE45595 (CAE45595) S-receptor kinase-like protein 2, complete
Length = 2946
Score = 157 bits (398), Expect = 6e-39
Identities = 102/297 (34%), Positives = 164/297 (54%), Gaps = 4/297 (1%)
Frame = +1
Query: 580 LRQVTGNFSEDNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVLSK 639
+ T +FS N LG GGFG VYKG L +G +IAVKR+ + + +G+ EF+ EI ++++
Sbjct: 1651 ISSATNHFSLSNKLGEGGFGPVYKGLLANGQEIAVKRLSNTS--GQGMEEFKNEIKLIAR 1824
Query: 640 VRHRHLVALLGHCINGNERLLVYEYMP---QGTLTQHLFEWRELGYTPLTWKQRVTVALD 696
++HR+LV L G ++ +E + M T ++ + W +R+ +
Sbjct: 1825 LQHRNLVKLFGCSVHQDENSHANKKMKILLDSTRSKLV-----------DWNKRLQIIDG 1971
Query: 697 VARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVET-RLAGT 755
+ARG+ YLH ++ IHRDLK SNILL D+M K++DFGL + + T R+ GT
Sbjct: 1972 IARGLLYLHQDSRLRIIHRDLKTSNILLDDEMNPKISDFGLARIFIGDQVEARTKRVMGT 2151
Query: 756 FGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHLVTWFRRVLINKE 815
+GY+ PEYA G + K DV++FGV+++E+I+G++ P +L++ R+ I +
Sbjct: 2152 YGYMPPEYAVHGSFSIKSDVFSFGVIVLEIISGKKIGRFYDPHHHLNLLSHAWRLWIEER 2331
Query: 816 NIPKAIDQTLNPDEETMESIYKVSELAGHCTAREPNQRPDMGHAVNVLVPMVEQWKP 872
+ + +D+ L+ D I + +A C R P RPDM V +L E KP
Sbjct: 2332 PL-ELVDELLD-DPVIPTEILRYIHVALLCVQRRPENRPDMLSIVLMLNGEKELPKP 2496
>BP032158
Length = 555
Score = 146 bits (368), Expect = 2e-35
Identities = 76/182 (41%), Positives = 113/182 (61%)
Frame = +2
Query: 576 SILVLRQVTGNFSEDNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEIT 635
S+ ++ T NF N +G GGFG VYKG L +G IAVK++ S + +G EF EI
Sbjct: 17 SLRQIKAATNNFDPANKIGEGGFGPVYKGVLSEGDVIAVKQLSSKS--KQGNREFINEIG 190
Query: 636 VLSKVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVAL 695
++S ++H +LV L G CI GN+ LLVYEYM +L + LF E L W+ R+ + +
Sbjct: 191 MISALQHPNLVKLYGCCIEGNQLLLVYEYMENNSLARALFGNEEQKLN-LNWRTRMKICV 367
Query: 696 DVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAGT 755
+A+G+ YLH ++ +HRD+K +N+LL D++AK++DFGL K + + TR+AGT
Sbjct: 368 GIAKGLAYLHEESRLKIVHRDIKATNVLLDKDLKAKISDFGLAKLDEEENTHISTRIAGT 547
Query: 756 FG 757
G
Sbjct: 548 IG 553
>TC16403 homologue to UP|Q9LKY4 (Q9LKY4) Pti1 kinase-like protein, partial
(55%)
Length = 876
Score = 145 bits (366), Expect = 3e-35
Identities = 79/166 (47%), Positives = 110/166 (65%), Gaps = 1/166 (0%)
Frame = +3
Query: 685 LTWKQRVTVALDVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDG 744
L+W QRV +A+ ARG+EYLH A+ IHR +K SNILL DD AK+ADF L APD
Sbjct: 36 LSWTQRVKIAVGAARGLEYLHEKAETHIIHRYIKSSNILLFDDDVAKIADFDLSNQAPDA 215
Query: 745 KYSVE-TRLAGTFGYLAPEYAATGRVTTKVDVYAFGVVLMELITGRRALDDSLPDERSHL 803
+ TR+ GTFGY APEYA TG++T+K DVY+FGVVL+EL+TGR+ +D +LP + L
Sbjct: 216 AARLHSTRVLGTFGYHAPEYAMTGQLTSKSDVYSFGVVLLELLTGRKPVDHTLPRGQQSL 395
Query: 804 VTWFRRVLINKENIPKAIDQTLNPDEETMESIYKVSELAGHCTARE 849
VTW L +++ + + +D L E ++S+ K++ +A C E
Sbjct: 396 VTWATPKL-SEDKVKQCVDARLK-GEYPVKSVAKMAAVAALCVQYE 527
>TC19726 similar to UP|Q9XIS3 (Q9XIS3) Lectin-like protein kinase, partial
(30%)
Length = 728
Score = 144 bits (363), Expect = 6e-35
Identities = 80/195 (41%), Positives = 117/195 (59%), Gaps = 4/195 (2%)
Frame = +3
Query: 580 LRQVTGNFSEDNILGRGGFGVVYKGEL-QDGTKIAVKRMESVAMGNKGLNEFQAEITVLS 638
L++ T NF E + LG+GG+GVVY+G L ++ ++AVK M K ++F +E+ +++
Sbjct: 147 LKKATNNFDEKHKLGQGGYGVVYRGMLPKEKLEVAVKMFSRDKM--KSTDDFLSELIIIN 320
Query: 639 KVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLF-EWRELGYTPLTWKQRVTVALDV 697
++RH+HLV L G C LLVY+YMP G+L H+F E TPL+W R + V
Sbjct: 321 RLRHKHLVRLQGWCHKNGVLLLVYDYMPNGSLDSHIFCEEGGTITTPLSWPLRYKIISGV 500
Query: 698 ARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGK--YSVETRLAGT 755
A + YLH+ Q +HRDLK SNI+L + AK+ DFGL + + K Y+ + GT
Sbjct: 501 ASALNYLHNEYDQKVVHRDLKASNIMLDSEFNAKLGDFGLARALENEKISYTELEGVHGT 680
Query: 756 FGYLAPEYAATGRVT 770
GY+APE TG+ T
Sbjct: 681 MGYIAPECFHTGKAT 725
>TC9461 similar to UP|O49840 (O49840) Protein kinase (AT2G02800/T20F6.6) ,
partial (37%)
Length = 484
Score = 143 bits (361), Expect = 1e-34
Identities = 77/163 (47%), Positives = 107/163 (65%), Gaps = 1/163 (0%)
Frame = +3
Query: 664 YMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDVARGVEYLHSLAQQSFIHRDLKPSNIL 723
+MP+G+L HLF G PL+W R+ VA+ ARG+ +LH+ A+ I+RD K SNIL
Sbjct: 3 FMPKGSLENHLFR---RGPQPLSWSVRMKVAIGAARGLSFLHN-AKSQVIYRDFKASNIL 170
Query: 724 LGDDMRAKVADFGLVKNAPDG-KYSVETRLAGTFGYLAPEYAATGRVTTKVDVYAFGVVL 782
L + AK++DFGL K P G + V T++ GT GY APEY ATGR+T K DVY+FGVVL
Sbjct: 171 LDAEFNAKLSDFGLAKAGPTGDRTHVSTQVMGTQGYAAPEYVATGRLTAKSDVYSFGVVL 350
Query: 783 MELITGRRALDDSLPDERSHLVTWFRRVLINKENIPKAIDQTL 825
+EL++GRRA+D ++ +LV W R L +K + + +D L
Sbjct: 351 LELLSGRRAVDKTIAGVDQNLVDWARPYLGDKRRLFRIMDSKL 479
>TC12493 similar to UP|Q9LP77 (Q9LP77) T1N15.9, partial (35%)
Length = 837
Score = 140 bits (354), Expect = 7e-34
Identities = 83/215 (38%), Positives = 121/215 (55%)
Frame = +3
Query: 592 ILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQAEITVLSKVRHRHLVALLGH 651
+LG+G FG YK ++ G +AVKR++ V EF+ +I + K+ H +LV L G+
Sbjct: 213 VLGKGTFGTTYKATMEMGRSVAVKRLKDVTATEM---EFREKIEEVGKLVHENLVPLRGY 383
Query: 652 CINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRVTVALDVARGVEYLHSLAQQS 711
+ +E+L+VY+YMP G+L+ L G TPL W+ R +AL A G+ YLHS S
Sbjct: 384 YFSRDEKLVVYDYMPMGSLSALLHANNGAGRTPLNWETRSAIALGAAHGIAYLHSQGPTS 563
Query: 712 FIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVETRLAGTFGYLAPEYAATGRVTT 771
H ++K SNILL +V+D GL A S R++ GY APE +V+
Sbjct: 564 -SHGNIKSSNILLTKSFEPRVSDLGLAYLAL--PTSTPNRIS---GYRAPEVTDARKVSQ 725
Query: 772 KVDVYAFGVVLMELITGRRALDDSLPDERSHLVTW 806
K DVY+FG++L+EL+TG+ SL +E L W
Sbjct: 726 KADVYSFGIMLLELLTGKPPTHSSLNEEGVDLPRW 830
>TC12343 similar to UP|CDK9_SCHPO (Q96WV9) Serine/threonine protein kinase
Cdk9 (Cyclin-dependent kinase Cdk9) , partial (7%)
Length = 723
Score = 140 bits (354), Expect = 7e-34
Identities = 77/179 (43%), Positives = 111/179 (61%)
Frame = +3
Query: 572 NSTMSILVLRQVTGNFSEDNILGRGGFGVVYKGELQDGTKIAVKRMESVAMGNKGLNEFQ 631
+ T S L T NF N LG GGFG V+KG+L DG +IAVK++ N+G +F
Sbjct: 198 HKTFSYETLVAATKNFHAVNKLGEGGFGPVFKGKLNDGREIAVKKLSR--RSNQGRTQFI 371
Query: 632 AEITVLSKVRHRHLVALLGHCINGNERLLVYEYMPQGTLTQHLFEWRELGYTPLTWKQRV 691
E +L++V+HR++V+L G+C +G+E+LLVYEY+P+ +L + LF R L WK+R
Sbjct: 372 NEAKLLTRVQHRNVVSLFGYCAHGSEKLLVYEYVPRESLDKLLF--RSQKKEQLDWKRRF 545
Query: 692 TVALDVARGVEYLHSLAQQSFIHRDLKPSNILLGDDMRAKVADFGLVKNAPDGKYSVET 750
+ VARG+ YLH + IHRD+K +NILL + K+ADFGL + P+ + V T
Sbjct: 546 DIISGVARGLLYLHEDSHDCIIHRDIKAANILLDEKWVPKIADFGLARIFPEDQTHVNT 722
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.133 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,762,171
Number of Sequences: 28460
Number of extensions: 219457
Number of successful extensions: 2179
Number of sequences better than 10.0: 408
Number of HSP's better than 10.0 without gapping: 1674
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1892
length of query: 935
length of database: 4,897,600
effective HSP length: 99
effective length of query: 836
effective length of database: 2,080,060
effective search space: 1738930160
effective search space used: 1738930160
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0108.8


