

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0108.10
(457 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
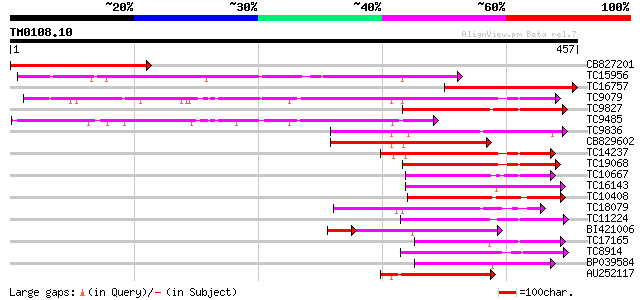
Score E
Sequences producing significant alignments: (bits) Value
CB827201 236 6e-63
TC15956 weakly similar to UP|LGT_CITUN (Q9MB73) Limonoid UDP-glu... 214 2e-56
TC16757 weakly similar to UP|Q9SBQ2 (Q9SBQ2) Anthocyanin 5-O-glu... 213 5e-56
TC9079 weakly similar to UP|Q8W4G1 (Q8W4G1) UDP-glucose glucosyl... 194 3e-50
TC9827 weakly similar to UP|Q8S9A2 (Q8S9A2) Glucosyltransferase-... 130 5e-31
TC9485 similar to UP|Q8S999 (Q8S999) Glucosyltransferase-10, par... 129 8e-31
TC9836 weakly similar to UP|Q9LXV0 (Q9LXV0) Glucosyltransferase-... 122 2e-28
CB829602 118 2e-27
TC14237 weakly similar to UP|AAR06913 (AAR06913) UDP-glycosyltra... 115 2e-26
TC19068 weakly similar to UP|AAR06913 (AAR06913) UDP-glycosyltra... 107 3e-24
TC10667 similar to UP|Q9M051 (Q9M051) Glucuronosyl transferase-l... 105 1e-23
TC16143 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltrans... 105 2e-23
TC10408 weakly similar to UP|Q9FUJ6 (Q9FUJ6) UDP-glucosyltransfe... 103 5e-23
TC18079 weakly similar to UP|UFO5_MANES (Q40287) Flavonol 3-O-gl... 101 2e-22
TC11224 weakly similar to UP|Q8S996 (Q8S996) Glucosyltransferase... 98 3e-21
BI421006 84 6e-21
TC17165 similar to UP|Q9SMG6 (Q9SMG6) Betanidin-5-O-glucosyltran... 94 4e-20
TC8914 similar to UP|Q9ZWS2 (Q9ZWS2) Flavonoid 3-O-galactosyl tr... 93 1e-19
BP039584 91 5e-19
AU252117 89 1e-18
>CB827201
Length = 374
Score = 236 bits (602), Expect = 6e-63
Identities = 114/114 (100%), Positives = 114/114 (100%)
Frame = +2
Query: 1 MPHHRFLLIIYPAQGHINPTLQFAKRLISSGAEHVTLCTTLHTYRRISNKPTLPNLTLLP 60
MPHHRFLLIIYPAQGHINPTLQFAKRLISSGAEHVTLCTTLHTYRRISNKPTLPNLTLLP
Sbjct: 32 MPHHRFLLIIYPAQGHINPTLQFAKRLISSGAEHVTLCTTLHTYRRISNKPTLPNLTLLP 211
Query: 61 FSDGFDDGFKLTPDTDYSLYATELKRRGTEFVTDLILSAAQEGKPFTCLIYTLL 114
FSDGFDDGFKLTPDTDYSLYATELKRRGTEFVTDLILSAAQEGKPFTCLIYTLL
Sbjct: 212 FSDGFDDGFKLTPDTDYSLYATELKRRGTEFVTDLILSAAQEGKPFTCLIYTLL 373
>TC15956 weakly similar to UP|LGT_CITUN (Q9MB73) Limonoid
UDP-glucosyltransferase (Limonoid glucosyltransferase)
(Limonoid GTase) (LGTase) , partial (65%)
Length = 1169
Score = 214 bits (545), Expect = 2e-56
Identities = 142/384 (36%), Positives = 201/384 (51%), Gaps = 25/384 (6%)
Frame = +2
Query: 7 LLIIYPAQGHINPTLQFAKRLISSGAEHVTLCTTLHTYRRISNKPTLPNLTLLPFSDG-- 64
LL+ +PAQGHINP L+ AK L + G+ VTL TT + + + + P SDG
Sbjct: 44 LLVSFPAQGHINPLLRLAKCLAAKGSS-VTLATTQTAGKSMRTATNITDKAPTPISDGSL 220
Query: 65 ----FDDGFKLTPDTD-----YSLYATELKRRGTEFVTDLILSAAQEGKPFTCLIYTLLL 115
FDDG L PD D SLY +L+ G + + +I A + +C+I L
Sbjct: 221 KFEFFDDG--LGPDDDSIRGNLSLYLQQLELVGRQQILKMIKEHADSNRQISCIINNPFL 394
Query: 116 HWAAEAARGLHLPTALLWVQPATVLDIIYYYFHGYHDCLLSK---IEFELPGLPFSLSPS 172
W + A +P+ALLW+Q + V Y YFH + + +LP L
Sbjct: 395 PWVVDVAEEQGIPSALLWIQSSAVFTAYYSYFHNLVTFPSKEDPLADVQLPSTSIVLKHC 574
Query: 173 DLPSFLLPS-NDSFIVSYFEEQFRELDVEANPTILVNSFEALEPEAMRAVEE-FNVIPIG 230
++P FL PS F+ + EQF+ L+ ILV+++E LE + + + + F + P+G
Sbjct: 575 EIPDFLHPSCTYPFLGTLILEQFKNLNKTF--CILVDTYEELEHDFIDYLSKTFLIRPVG 748
Query: 231 PLIPSAFLDGKDLNDTSFGGDIFRLSNGCAEWLDLQAEKSVVYVSFGSFVVLSKTQMEEI 290
PL S+ + GD F S+ C EWL+ + + SVVYVSFGS V L + Q++EI
Sbjct: 749 PLFKSS-----QAKSAAIRGD-FMKSDDCIEWLNSKPQSSVVYVSFGSIVYLPQEQVDEI 910
Query: 291 ARALLDCGHPFLWVIREMEEELSCR---------EELEEKGKVVTWCSQVEVLSHRSVGC 341
A L FLWV++ E + EE E+G+VV W Q EVL+H SV C
Sbjct: 911 AFGLKKSQVSFLWVMKPPPEVTGLQPHVLPYGFLEETGERGRVVQWSPQEEVLAHPSVSC 1090
Query: 342 FLTHCGWNSTMESLVSGVPMVAFP 365
FLTHCGWNS+ME+L SGVP++ FP
Sbjct: 1091FLTHCGWNSSMEALTSGVPVLTFP 1162
>TC16757 weakly similar to UP|Q9SBQ2 (Q9SBQ2) Anthocyanin
5-O-glucosyltransferase, partial (19%)
Length = 638
Score = 213 bits (542), Expect = 5e-56
Identities = 105/107 (98%), Positives = 105/107 (98%)
Frame = +2
Query: 351 TMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGVRMDHSVNEDGVVEGGEIRKCLDAVM 410
TMESL SGVPM AFPRFSDQMTNAKMVEDVWKIGVRMDHSVNEDGVVEGGEIRKCLDAVM
Sbjct: 2 TMESLXSGVPMXAFPRFSDQMTNAKMVEDVWKIGVRMDHSVNEDGVVEGGEIRKCLDAVM 181
Query: 411 GNGEKAEEVRRNAEKFKGLAMEAGKEGGSSEKNLMAFLAVGGYGSCN 457
GNGEKAEEVRRNAEKFKGLAMEAGKEGGSSEKNLMAFLAVGGYGSCN
Sbjct: 182 GNGEKAEEVRRNAEKFKGLAMEAGKEGGSSEKNLMAFLAVGGYGSCN 322
>TC9079 weakly similar to UP|Q8W4G1 (Q8W4G1) UDP-glucose
glucosyltransferase, partial (65%)
Length = 1585
Score = 194 bits (493), Expect = 3e-50
Identities = 152/471 (32%), Positives = 238/471 (50%), Gaps = 38/471 (8%)
Frame = +1
Query: 12 PAQGHINPTLQFAKRLISSGAEHVTLCTTLHTYRRI--SNKPT----LPNLTLLPFSDGF 65
P QGHINP Q AK L G H+T T + ++R+ S P LP+ DG
Sbjct: 40 PLQGHINPLFQLAKLLHLRGF-HITFVHTEYNHKRLIKSRGPNALDGLPDFVFETIPDGL 216
Query: 66 DDGFKLTPDTDYSLYATELKRRGTEFVTDLILSAAQEGK-----PFTCLIYTLLLHWAAE 120
+DG YSL + + F DL+ + P TCL+ LL+ + +
Sbjct: 217 EDGDGEVSQDLYSLCDSIRNKCHLPF-QDLLAKLHRSASAGLVPPVTCLVSDLLMTFTIQ 393
Query: 121 AARGLHLPTALLWVQPA----------TVLD--II-----YYYFHGYHDCLLSKIEFELP 163
AA+ L LP LL A T+LD +I Y +GY D +K+++ +P
Sbjct: 394 AAQQLQLPILLLCPASASTFMSFLHFQTLLDKGVIPLKDESYLTNGYLD---TKVDW-IP 561
Query: 164 GLPFSLSPSDLPSFLLPSNDSFIVSYFEEQFRELDVEANPTILVNSFEALEPEAMRAVEE 223
G+ + DLP F+ ++ + I+ F + + A+ I+ N+F LE + + A+
Sbjct: 562 GMQ-NFRLKDLPDFIRTTDPNDIMLEFMVEVADRAKRAS-AIVFNTFNELERDVLSALST 735
Query: 224 F--NVIPIGPLIPSAFLDGKDLND-TSFGGDIFRLSNGCAEWLDLQAEKSVVYVSFGSFV 280
++ PIGP +FL+ N S G ++++ C +WL+ + SVVYV+FGS
Sbjct: 736 MLPSLYPIGPF--PSFLNQAPQNQLASLGSNLWKEDTKCLQWLESKEPGSVVYVNFGSIT 909
Query: 281 VLSKTQMEEIARALLDCGHPFLWVIR-----EMEEELSCR--EELEEKGKVVTWCSQVEV 333
V+S Q+ E A L + PFLW+IR + LS + +E +G + +WC Q +V
Sbjct: 910 VMSPEQLLEFAWGLANGKKPFLWIIRPDLVADGSAILSPKFVDETSNRGLIASWCPQEKV 1089
Query: 334 LSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGVRMDHSVNE 393
L+H SVG FLTHCGWNST+ES+ +GVPM+ +P +DQ TN + + + W IG+ +D +V
Sbjct: 1090LNHPSVGGFLTHCGWNSTIESICAGVPMLCWPFVADQPTNCRYICNEWDIGIEVDTNVKR 1269
Query: 394 DGVVEGGEIRKCLDAVMGNGEKAEEVRRNAEKFKGLAMEAGKEGGSSEKNL 444
+ E+ + +M G+K +++R+ + K A E + GG S NL
Sbjct: 1270E------EVENLIHELM-VGDKGKKMRQRTMELKKKAEEDTRPGGCSYMNL 1401
>TC9827 weakly similar to UP|Q8S9A2 (Q8S9A2) Glucosyltransferase-7
(Fragment), partial (42%)
Length = 611
Score = 130 bits (327), Expect = 5e-31
Identities = 62/133 (46%), Positives = 90/133 (67%)
Frame = +2
Query: 317 ELEEKGKVVTWCSQVEVLSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKM 376
E EKG +VTWC Q+ VL H+++GCFLTHCGWNST+ES+ GVP++A P ++D +TN K+
Sbjct: 11 ETTEKGLIVTWCPQLLVLKHQALGCFLTHCGWNSTLESVSLGVPVIAMPLWTDPVTNGKL 190
Query: 377 VEDVWKIGVRMDHSVNEDGVVEGGEIRKCLDAVMGNGEKAEEVRRNAEKFKGLAMEAGKE 436
+ DVWK GVR +E +V I+ C+ ++ EK E++ NA K+ LA ++ E
Sbjct: 191 IVDVWKTGVRA--VADEKEIVRRETIKNCIKEII-ETEKINEIKSNAIKWMNLAKDSLAE 361
Query: 437 GGSSEKNLMAFLA 449
GG S+KN+ F+A
Sbjct: 362 GGRSDKNIAEFVA 400
>TC9485 similar to UP|Q8S999 (Q8S999) Glucosyltransferase-10, partial (75%)
Length = 1195
Score = 129 bits (325), Expect = 8e-31
Identities = 119/385 (30%), Positives = 177/385 (45%), Gaps = 41/385 (10%)
Frame = +1
Query: 2 PHHRFLLIIYPAQGHINPTLQFAKRLISSGAEHVTLCTTLHTYRRISNKPTLPNLTLLPF 61
PH + I YPAQGHINP L+ AK L G HVT T + ++R+ +L LP
Sbjct: 88 PH--VVCIPYPAQGHINPMLKLAKLLHFKGGFHVTFVNTEYNHKRLLKSRGPDSLNGLPS 261
Query: 62 S--DGFDDGFKLTPDTDY-------SLYATELKRRGTEF--VTDLILSAAQEGKPFTCLI 110
+ DG P+TD SL + K F + + + + P TC++
Sbjct: 262 FRFETIPDGL---PETDVDVTQDIPSLCISTRKTCLPHFKKLLSKLNDVSSDVPPVTCIV 432
Query: 111 YTLLLHWAAEAARGLHLPTALLWVQPAT-VLDIIYY----------------YFHGYHDC 153
+ + +AA L++P L W A + + Y +GY
Sbjct: 433 SDGCMSFTLDAAIELNIPEVLFWTTSACGFMGYVQYRELIEKGIIPLKDSSDITNGY--- 603
Query: 154 LLSKIEFELPGLPFSLSPSDLPSFLLPS--NDSFIVSYFEEQFRELDVEANPTILVNSFE 211
L + IE+ LPG+ ++ DLPSFL + ND + E R L A I++N+F+
Sbjct: 604 LETTIEW-LPGMK-NIRLKDLPSFLRTTDPNDKMLDFLTGECQRALKASA---IILNTFD 768
Query: 212 ALEPEAMRAVEEF--NVIPIGPLIPSAFLDGKDLNDTSFGGDIFRLSNGCAEWLDLQAEK 269
ALE + + A V IGPL D D N S G ++++ + C +WLD +
Sbjct: 769 ALEHDVLEAFSSILPPVYSIGPL-HLLIKDVTDKNLNSLGSNLWKEDSECLKWLDTKEPN 945
Query: 270 SVVYVSFGSFVVLSKTQMEEIARALLDCGHPFLWVIRE---------MEEELSCREELEE 320
SVVYV+FGS V++ QM E A L + FLWVIR + EE +
Sbjct: 946 SVVYVNFGSITVMTSEQMVEFAWGLANSNKTFLWVIRPDLVAGKHAVLPEEFVA--ATND 1119
Query: 321 KGKVVTWCSQVEVLSHRSVGCFLTH 345
+G++ +W Q +VL+H ++G FLTH
Sbjct: 1120RGRLSSWTPQEDVLTHPAIGGFLTH 1194
>TC9836 weakly similar to UP|Q9LXV0 (Q9LXV0) Glucosyltransferase-like
protein, partial (55%)
Length = 1286
Score = 122 bits (305), Expect = 2e-28
Identities = 73/209 (34%), Positives = 118/209 (55%), Gaps = 18/209 (8%)
Frame = +1
Query: 259 CAEWLDLQAEKSVVYVSFGSFVVLSKTQMEEIARALLDCGHPFLWVIR-----EMEEELS 313
C +WLD + SV++VSFGS +S +QM ++A AL G F+WV+R ++ E
Sbjct: 379 CRKWLDTKPSNSVLFVSFGSMNTISASQMMQLATALDRSGRNFIWVVRPPIGFDINSEFR 558
Query: 314 CREELEE--------KGKVV-TWCSQVEVLSHRSVGCFLTHCGWNSTMESLVSGVPMVAF 364
E L E KG VV W QVE+LSH +V FL+HCGWNS +ESL GVP++ +
Sbjct: 559 AEEWLPEGFLGRVAKKGLVVHDWAPQVEILSHGAVSAFLSHCGWNSVLESLSHGVPILGW 738
Query: 365 PRFSDQMTNAKMVEDVWKIGVRMDHSVNEDGVVEGGEIRKCLDAVMGNGEKAEEVRRNAE 424
P ++Q N KM+E+ ++GV ++ + + V ++ + ++ VM E ++R+NA
Sbjct: 739 PMAAEQFFNCKMLEE--ELGVCVEVARGKRCEVRHEDLVEKIELVMNEAESGVKIRKNAG 912
Query: 425 KFKGLAMEAGKE----GGSSEKNLMAFLA 449
+ + +A ++ GSS + + FL+
Sbjct: 913 NIREMIRDAVRDEDGYKGSSVRAIDEFLS 999
>CB829602
Length = 542
Score = 118 bits (296), Expect = 2e-27
Identities = 56/137 (40%), Positives = 87/137 (62%), Gaps = 7/137 (5%)
Frame = +1
Query: 259 CAEWLDLQAEKSVVYVSFGSFVVLSKTQMEEIARALLDCGHPFLWVIR----EMEEELSC 314
C +WLD Q SV+YV+FGS + ++ Q+ E+A + + F+WVIR E E +
Sbjct: 94 CLKWLDSQEPSSVLYVNFGSVINMTPQQLVELAWGIANSKKKFIWVIRPDLVEGEASIVL 273
Query: 315 RE---ELEEKGKVVTWCSQVEVLSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQM 371
E E +++G +++WC Q +VL H ++G FLTHCGWNST+ES+ SGVP++ P F+DQ
Sbjct: 274 PEIVAETKDRGIMLSWCPQEQVLKHSALGGFLTHCGWNSTIESISSGVPLICSPFFNDQF 453
Query: 372 TNAKMVEDVWKIGVRMD 388
N++ + W G+ M+
Sbjct: 454 VNSRYICSEWDFGMEMN 504
>TC14237 weakly similar to UP|AAR06913 (AAR06913) UDP-glycosyltransferase
85A8, partial (27%)
Length = 999
Score = 115 bits (288), Expect = 2e-26
Identities = 59/149 (39%), Positives = 94/149 (62%), Gaps = 8/149 (5%)
Frame = +2
Query: 300 PFLWVIREM----EEELSCREE----LEEKGKVVTWCSQVEVLSHRSVGCFLTHCGWNST 351
PFLW++R E+ ++ +E ++ +G + +WC Q EVLSH S+G FLTHCGWNST
Sbjct: 215 PFLWILRPDVVMGEDSINVPQEFLDEIKGRGYIASWCFQEEVLSHPSIGAFLTHCGWNST 394
Query: 352 MESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGVRMDHSVNEDGVVEGGEIRKCLDAVMG 411
+E + +G+P++ +P FS+Q TN++ W+IG+ ++H V D EI ++ +M
Sbjct: 395 IEGISAGLPLICWPFFSEQHTNSRYACTTWEIGMEVNHDVKRD------EITTLVNEMM- 553
Query: 412 NGEKAEEVRRNAEKFKGLAMEAGKEGGSS 440
GEK +E+R+ ++K A+EA GGSS
Sbjct: 554 KGEKGKEMRKKGLEWKKKAIEATGLGGSS 640
>TC19068 weakly similar to UP|AAR06913 (AAR06913) UDP-glycosyltransferase
85A8, partial (26%)
Length = 565
Score = 107 bits (268), Expect = 3e-24
Identities = 52/128 (40%), Positives = 83/128 (64%)
Frame = +1
Query: 317 ELEEKGKVVTWCSQVEVLSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKM 376
E+ +G + +WCSQ VL+H S+G FLTHCGWNST+ES+ +GVPM+ +P F+DQ+TN +
Sbjct: 37 EISSRGLIASWCSQEHVLNHPSIGGFLTHCGWNSTIESISAGVPMLCWPFFADQLTNCRS 216
Query: 377 VEDVWKIGVRMDHSVNEDGVVEGGEIRKCLDAVMGNGEKAEEVRRNAEKFKGLAMEAGKE 436
+ + W IG+++D + + E+ K ++ +M GEK +++R+ + K A E +
Sbjct: 217 ICNEWDIGMQIDTNGKRE------EVEKLINELM-VGEKGKKMRQKTMELKKRAEEDTRP 375
Query: 437 GGSSEKNL 444
GG S NL
Sbjct: 376 GGCSYMNL 399
>TC10667 similar to UP|Q9M051 (Q9M051) Glucuronosyl transferase-like
protein, partial (20%)
Length = 824
Score = 105 bits (263), Expect = 1e-23
Identities = 58/121 (47%), Positives = 73/121 (59%)
Frame = +1
Query: 320 EKGKVVTWCSQVEVLSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKMVED 379
E G V W Q +VL H ++G F THCGWNST+ES+ GVPM+ P F DQ NAK V D
Sbjct: 1 EFGTSVKWAPQEQVLKHPAIGAFWTHCGWNSTLESVCEGVPMICSPCFGDQKVNAKYVSD 180
Query: 380 VWKIGVRMDHSVNEDGVVEGGEIRKCLDAVMGNGEKAEEVRRNAEKFKGLAMEAGKEGGS 439
VWK+GV++ N+ G GEI K + +M G++A E+R N K K A EGGS
Sbjct: 181 VWKVGVQLQ---NKPG---RGEIEKTISLLM-LGDEAYEIRGNILKLKEKANVCLSEGGS 339
Query: 440 S 440
S
Sbjct: 340 S 342
>TC16143 similar to UP|Q7XZD0 (Q7XZD0) Isoflavonoid glucosyltransferase,
partial (30%)
Length = 604
Score = 105 bits (262), Expect = 2e-23
Identities = 53/137 (38%), Positives = 83/137 (59%), Gaps = 8/137 (5%)
Frame = +1
Query: 320 EKGKVVT-WCSQVEVLSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKMVE 378
EKG ++ W QV +L HR+VG F+THCGWNST+E++ +GVPM+ +P +Q N K++
Sbjct: 4 EKGMIIRGWVPQVVILGHRAVGAFVTHCGWNSTVEAVSAGVPMITWPVVGEQFYNEKLIT 183
Query: 379 DVWKIGVRMDHSV-------NEDGVVEGGEIRKCLDAVMGNGEKAEEVRRNAEKFKGLAM 431
+V IGV + + +V I K + +M G++ E++RR A+++ A
Sbjct: 184 EVRGIGVEVGAEECCLMGFDKREKLVSRESIEKAVRRLMDGGDEGEQIRRRAQEYGEKAR 363
Query: 432 EAGKEGGSSEKNLMAFL 448
+A +EGGSS KNL A +
Sbjct: 364 QAVEEGGSSHKNLTALI 414
>TC10408 weakly similar to UP|Q9FUJ6 (Q9FUJ6) UDP-glucosyltransferase HRA25,
partial (19%)
Length = 571
Score = 103 bits (258), Expect = 5e-23
Identities = 50/128 (39%), Positives = 80/128 (62%)
Frame = +1
Query: 321 KGKVVTWCSQVEVLSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKMVEDV 380
KGKVV W Q ++L+H ++ CF++HCGWNST+E + + VP + +P +DQ N + DV
Sbjct: 13 KGKVVKWGPQKKILNHPAIACFISHCGWNSTIEGVHASVPFLCWPFSTDQFLNKSFICDV 192
Query: 381 WKIGVRMDHSVNEDGVVEGGEIRKCLDAVMGNGEKAEEVRRNAEKFKGLAMEAGKEGGSS 440
WKIG+ +D +E+G++ GEIR ++ V+ N E+++ + K K + + EGG S
Sbjct: 193 WKIGLGLDK--DENGIIPKGEIRTKVEQVIVN----EDLKARSLKLKEITINNLAEGGQS 354
Query: 441 EKNLMAFL 448
NL F+
Sbjct: 355 SNNLKKFI 378
>TC18079 weakly similar to UP|UFO5_MANES (Q40287) Flavonol
3-O-glucosyltransferase 5 (UDP-glucose flavonoid
3-O-glucosyltransferase 5) , partial (33%)
Length = 889
Score = 101 bits (252), Expect = 2e-22
Identities = 71/191 (37%), Positives = 98/191 (51%), Gaps = 20/191 (10%)
Frame = +3
Query: 262 WLDLQAEKSVVYVSFGSFVVLSKTQMEEIARALLDCGHPFLWVIREMEE----------- 310
WLD Q SV+YVSFGS + + QM EIA L H F+WV+R E
Sbjct: 69 WLDKQPAGSVIYVSFGSGGTMPQGQMTEIALGLELSQHRFVWVVRPPNEADASATFFGFG 248
Query: 311 -ELSCR-------EELEEKGKVVT-WCSQVEVLSHRSVGCFLTHCGWNSTMESLVSGVPM 361
E++ E G VV W Q E+L H + G F+T CGWNS +ES+++GVPM
Sbjct: 249 NEMALNYLPEGFMNRTREFGVVVPMWAPQAEILGHPATGGFVTPCGWNSVLESVLNGVPM 428
Query: 362 VAFPRFSDQMTNAKMVEDVWKIGVRMDHSVNEDGVVEGGEIRKCLDAVMGNGEKAEEVRR 421
VA+P +S+Q NA M+ + ++GV + V E GVV C D V+ + VRR
Sbjct: 429 VAWPLYSEQKMNAYMLSE--ELGVAVRVKVAEGGVV-------CRDQVV------DMVRR 563
Query: 422 NAEKFKGLAME 432
+G+ M+
Sbjct: 564 VMVSEEGMGMK 596
>TC11224 weakly similar to UP|Q8S996 (Q8S996) Glucosyltransferase-13
(Fragment), partial (19%)
Length = 596
Score = 97.8 bits (242), Expect = 3e-21
Identities = 53/135 (39%), Positives = 79/135 (58%)
Frame = +3
Query: 316 EELEEKGKVVTWCSQVEVLSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAK 375
E + +GKVV+W Q+E+L H SVG FLTH GWNS +E +V GVPM+ P F DQ N
Sbjct: 15 ERTKSQGKVVSWAPQMEILKHASVGVFLTHGGWNSVLECIVGGVPMIGRPFFGDQRINIW 194
Query: 376 MVEDVWKIGVRMDHSVNEDGVVEGGEIRKCLDAVMGNGEKAEEVRRNAEKFKGLAMEAGK 435
M+ + +GV + ++GV+ I K L + M GE+ + +R+ + K +A +A +
Sbjct: 195 MLAKLLGVGVGL-----QNGVLAKETILKTLKSTM-TGEEGKVMRQKMAELKEMAWKAVE 356
Query: 436 EGGSSEKNLMAFLAV 450
GSS KNL + +
Sbjct: 357 PDGSSTKNLCTLMQI 401
>BI421006
Length = 519
Score = 83.6 bits (205), Expect(2) = 6e-21
Identities = 44/137 (32%), Positives = 74/137 (53%), Gaps = 15/137 (10%)
Frame = +2
Query: 276 FGSFVVLSKTQMEEIARALLDCGHPFLWVIREMEE-ELSCREELEEKGK----------- 323
F F + Q+E+IA L F+WV+R+ ++ ++ ++++E+G
Sbjct: 101 FWFFSTFIEEQIEQIANGLEQSKQKFIWVLRDADKGDIFDGDKVKERGLPKGFEKRVEGM 280
Query: 324 ---VVTWCSQVEVLSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKMVEDV 380
V W Q+E+LSH S G F++HCGWNS +ES+ GVP+ A+P SDQ N ++ +V
Sbjct: 281 GLVVRDWAPQLEILSHPSTGGFMSHCGWNSCIESMSMGVPIAAWPMHSDQPRNTVLITEV 460
Query: 381 WKIGVRMDHSVNEDGVV 397
K+ + + D +V
Sbjct: 461 LKVALVVQDXAQRDELV 511
Score = 33.9 bits (76), Expect(2) = 6e-21
Identities = 15/23 (65%), Positives = 17/23 (73%)
Frame = +3
Query: 257 NGCAEWLDLQAEKSVVYVSFGSF 279
N EWLD Q +KSV+YVSFG F
Sbjct: 45 NFVMEWLDRQEQKSVLYVSFGFF 113
>TC17165 similar to UP|Q9SMG6 (Q9SMG6) Betanidin-5-O-glucosyltransferase,
partial (24%)
Length = 546
Score = 94.4 bits (233), Expect = 4e-20
Identities = 51/127 (40%), Positives = 74/127 (58%), Gaps = 5/127 (3%)
Frame = +1
Query: 327 WCSQVEVLSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGV- 385
W QV +L H ++G F+THCGWNST+E + +GVPMV +P ++Q N K+V +V KIGV
Sbjct: 13 WAPQVLILEHEAIGAFVTHCGWNSTLEGVAAGVPMVTWPIAAEQFYNEKLVTEVLKIGVP 192
Query: 386 ----RMDHSVNEDGVVEGGEIRKCLDAVMGNGEKAEEVRRNAEKFKGLAMEAGKEGGSSE 441
+ V E V+ + K + +M GE+AEE+R + A A +EGGSS
Sbjct: 193 VGAKKWGRMVLEGDGVKSDAVEKGVKRIM-EGEEAEEMRNRVKVLSQQAQWAVEEGGSSY 369
Query: 442 KNLMAFL 448
+L A +
Sbjct: 370 SDLNALI 390
>TC8914 similar to UP|Q9ZWS2 (Q9ZWS2) Flavonoid 3-O-galactosyl transferase,
partial (32%)
Length = 627
Score = 92.8 bits (229), Expect = 1e-19
Identities = 46/136 (33%), Positives = 78/136 (56%), Gaps = 1/136 (0%)
Frame = +2
Query: 316 EELEEKGKVVTWCSQVEVLSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAK 375
E + GK+V W Q ++L H SVG F+THCG NS ES+ +GVPM+ P F D +
Sbjct: 47 ERTKNSGKIVPWAPQTQILGHESVGVFVTHCGCNSVFESISNGVPMICRPFFGDHGMAGR 226
Query: 376 MVEDVWKIGVRMDHSV-NEDGVVEGGEIRKCLDAVMGNGEKAEEVRRNAEKFKGLAMEAG 434
++ED+W+IGV+++ V +DG+V K L+ ++ E+ + +R A+K K ++A
Sbjct: 227 IIEDIWEIGVKIEGGVFTQDGLV------KSLNLILVQ-EEGKIMREKAQKVKKTVLDAA 385
Query: 435 KEGGSSEKNLMAFLAV 450
G + ++ + +
Sbjct: 386 GPQGKATQDFKTLVGI 433
>BP039584
Length = 545
Score = 90.5 bits (223), Expect = 5e-19
Identities = 44/120 (36%), Positives = 72/120 (59%), Gaps = 6/120 (5%)
Frame = -3
Query: 327 WCSQVEVLSHRSVGCFLTHCGWNSTMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGVR 386
W Q+ +L H ++G +THCGWN+T+ES+++G+PM P F++Q N K++ DV +GV
Sbjct: 537 WAPQLLILEHPAIGGVVTHCGWNTTLESVIAGLPMATMPLFAEQFYNEKLLVDVLGVGVS 358
Query: 387 M------DHSVNEDGVVEGGEIRKCLDAVMGNGEKAEEVRRNAEKFKGLAMEAGKEGGSS 440
+ + +V D +V+ I K + +MG GE+A E+RR + A + + GGSS
Sbjct: 357 IGMKKWRNWNVVGDEIVKRENIVKAISLLMGGGEEALEMRRRVRELSDAAKKTIQPGGSS 178
>AU252117
Length = 350
Score = 89.4 bits (220), Expect = 1e-18
Identities = 39/102 (38%), Positives = 64/102 (62%), Gaps = 10/102 (9%)
Frame = +3
Query: 300 PFLWVIR----------EMEEELSCREELEEKGKVVTWCSQVEVLSHRSVGCFLTHCGWN 349
PFLW++R + +E +E++++G + +WC+Q +VL+H S+G FLTHC WN
Sbjct: 51 PFLWIMRPDVVMGGEFINLPQEFL--DEIKDRGYIASWCNQEQVLAHPSIGVFLTHCDWN 224
Query: 350 STMESLVSGVPMVAFPRFSDQMTNAKMVEDVWKIGVRMDHSV 391
ST+ + SGVPM+ +P F++Q TN + W G+ ++H V
Sbjct: 225 STVVCVSSGVPMICWPFFAEQQTNCRYACTTWGTGMDINHDV 350
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.321 0.138 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,519,481
Number of Sequences: 28460
Number of extensions: 126568
Number of successful extensions: 711
Number of sequences better than 10.0: 102
Number of HSP's better than 10.0 without gapping: 678
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 682
length of query: 457
length of database: 4,897,600
effective HSP length: 93
effective length of query: 364
effective length of database: 2,250,820
effective search space: 819298480
effective search space used: 819298480
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0108.10


