

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
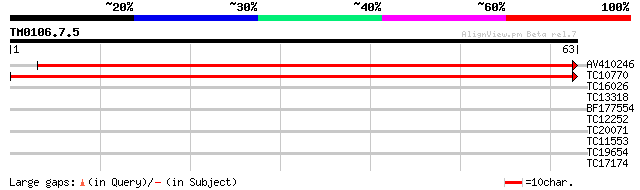
Query= TM0106.7.5
(63 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AV410246 122 1e-29
TC10770 weakly similar to UP|RN12_HUMAN (Q9NVW2) RING finger pro... 115 2e-27
TC16026 similar to UP|Q9SGZ3 (Q9SGZ3) F28K19.29, partial (32%) 27 0.60
TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (... 26 1.7
BF177554 24 5.1
TC12252 weakly similar to GB|AAB66486.1|2318131|AF014824 histone... 24 5.1
TC20071 weakly similar to UP|Q41326 (Q41326) Peroxidase (Fragmen... 24 6.6
TC11553 weakly similar to UP|BAD10712 (BAD10712) Proline-rich pr... 24 6.6
TC19654 24 6.6
TC17174 weakly similar to UP|Q940N9 (Q940N9) AT4g21910/T8O5_120,... 23 8.6
>AV410246
Length = 426
Score = 122 bits (307), Expect = 1e-29
Identities = 60/60 (100%), Positives = 60/60 (100%)
Frame = +3
Query: 4 NDLEENPDLRFNLSLYQNIEFQPSEMASVTDGEELPSVPLEELLADLELGEDEDEEDFMM 63
NDLEENPDLRFNLSLYQNIEFQPSEMASVTDGEELPSVPLEELLADLELGEDEDEEDFMM
Sbjct: 3 NDLEENPDLRFNLSLYQNIEFQPSEMASVTDGEELPSVPLEELLADLELGEDEDEEDFMM 182
>TC10770 weakly similar to UP|RN12_HUMAN (Q9NVW2) RING finger protein 12
(LIM domain interacting RING finger protein) (RING
finger LIM domain-binding protein) (R-LIM) (NY-REN-43
antigen), partial (4%)
Length = 481
Score = 115 bits (287), Expect = 2e-27
Identities = 56/63 (88%), Positives = 60/63 (94%)
Frame = +2
Query: 1 QFLNDLEENPDLRFNLSLYQNIEFQPSEMASVTDGEELPSVPLEELLADLELGEDEDEED 60
QFL DLEENPDLRFN+SLY+N E+QPSE+ASVTDGEELPSVPLEELLADLELGEDEDEED
Sbjct: 98 QFLKDLEENPDLRFNISLYRNKEYQPSEIASVTDGEELPSVPLEELLADLELGEDEDEED 277
Query: 61 FMM 63
MM
Sbjct: 278 DMM 286
>TC16026 similar to UP|Q9SGZ3 (Q9SGZ3) F28K19.29, partial (32%)
Length = 1183
Score = 27.3 bits (59), Expect = 0.60
Identities = 18/38 (47%), Positives = 22/38 (57%), Gaps = 2/38 (5%)
Frame = -2
Query: 25 QPSEMASVTDGEELPSVPLEELLADL--ELGEDEDEED 60
QPSE VT+GEE + EE D E GE E+EE+
Sbjct: 771 QPSEKKGVTEGEE*KELGGEEQGRDEFDEDGEGEEEEE 658
>TC13318 similar to UP|CAA20314 (CAA20314) SPBC30B4.01c protein (SPBC3D6.14C
protein) (Fragment), partial (15%)
Length = 543
Score = 25.8 bits (55), Expect = 1.7
Identities = 18/47 (38%), Positives = 24/47 (50%), Gaps = 1/47 (2%)
Frame = +3
Query: 15 NLSLYQNIEFQPSEMA-SVTDGEELPSVPLEELLADLELGEDEDEED 60
+L LY+ IE A V D E P P + + D E ED+DEE+
Sbjct: 141 DLRLYRLIELHNGLQAWLVHDPEIYPDGPPKPVQTDNEEEEDDDEEE 281
>BF177554
Length = 246
Score = 24.3 bits (51), Expect = 5.1
Identities = 15/38 (39%), Positives = 18/38 (46%)
Frame = +1
Query: 23 EFQPSEMASVTDGEELPSVPLEELLADLELGEDEDEED 60
EF SE EE EE + E GE+E+EED
Sbjct: 1 EFGTSEQQDEDSCEEEAGESSEEEEEEEEEGEEEEEED 114
>TC12252 weakly similar to GB|AAB66486.1|2318131|AF014824 histone
deacetylase {Arabidopsis thaliana;} , partial (12%)
Length = 559
Score = 24.3 bits (51), Expect = 5.1
Identities = 12/26 (46%), Positives = 16/26 (61%), Gaps = 1/26 (3%)
Frame = +1
Query: 36 EELPSVPLEELLADLELGE-DEDEED 60
+ PSV +E D +LGE DED +D
Sbjct: 16 QHAPSVQFQERPPDYDLGEADEDHDD 93
>TC20071 weakly similar to UP|Q41326 (Q41326) Peroxidase (Fragment), partial
(92%)
Length = 535
Score = 23.9 bits (50), Expect = 6.6
Identities = 12/22 (54%), Positives = 13/22 (58%)
Frame = +1
Query: 34 DGEELPSVPLEELLADLELGED 55
D EELP PLEE L+ G D
Sbjct: 355 D*EELPETPLEETSKMLKTGLD 420
>TC11553 weakly similar to UP|BAD10712 (BAD10712) Proline-rich protein
family-like, partial (27%)
Length = 493
Score = 23.9 bits (50), Expect = 6.6
Identities = 15/37 (40%), Positives = 20/37 (53%), Gaps = 1/37 (2%)
Frame = -1
Query: 25 QPSEMASVTDGEELPSVPLE-ELLADLELGEDEDEED 60
+P + S+ D E +P+ LA L G DEDEED
Sbjct: 196 RPWQYFSLMDDLERGPLPISAHPLASLPPGVDEDEED 86
>TC19654
Length = 530
Score = 23.9 bits (50), Expect = 6.6
Identities = 16/44 (36%), Positives = 21/44 (47%), Gaps = 1/44 (2%)
Frame = -2
Query: 18 LYQNIEFQPSEMASVTD-GEELPSVPLEELLADLELGEDEDEED 60
L + E +AS +D G E+P V E +L G DED D
Sbjct: 274 LLTSFEIVSELLASFSDMGNEIPLVDSELMLRLASGGNDEDAAD 143
>TC17174 weakly similar to UP|Q940N9 (Q940N9) AT4g21910/T8O5_120, partial
(18%)
Length = 723
Score = 23.5 bits (49), Expect = 8.6
Identities = 11/25 (44%), Positives = 15/25 (60%)
Frame = +2
Query: 6 LEENPDLRFNLSLYQNIEFQPSEMA 30
+E LR+NL +YQ I F+P A
Sbjct: 269 IELTGGLRWNLIIYQIISFKPRPSA 343
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.310 0.135 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 770,920
Number of Sequences: 28460
Number of extensions: 7142
Number of successful extensions: 42
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 42
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 42
length of query: 63
length of database: 4,897,600
effective HSP length: 39
effective length of query: 24
effective length of database: 3,787,660
effective search space: 90903840
effective search space used: 90903840
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 49 (23.5 bits)
Lotus: description of TM0106.7.5


