

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
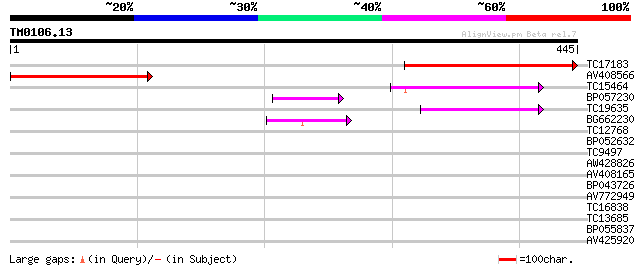
Query= TM0106.13
(445 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC17183 weakly similar to UP|Q95RC5 (Q95RC5) LD44506p (CG5745-PA... 284 2e-77
AV408566 225 1e-59
TC15464 54 7e-08
BP057230 45 2e-05
TC19635 45 2e-05
BG662230 43 9e-05
TC12768 similar to UP|Q9FH10 (Q9FH10) Serine/threonine protein k... 30 1.0
BP052632 28 4.0
TC9497 28 4.0
AW428826 27 5.2
AV408165 27 5.2
BP043726 27 5.2
AV772949 27 6.8
TC16838 similar to UP|BAD07764 (BAD07764) GTPase activating prot... 27 6.8
TC13685 similar to UP|Q9FJH5 (Q9FJH5) Gb|AAC98457.1 (At5g60800),... 27 6.8
BP055837 27 6.8
AV425920 27 8.9
>TC17183 weakly similar to UP|Q95RC5 (Q95RC5) LD44506p (CG5745-PA), partial
(6%)
Length = 678
Score = 284 bits (726), Expect = 2e-77
Identities = 135/135 (100%), Positives = 135/135 (100%)
Frame = +1
Query: 311 QPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDT 370
QPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDT
Sbjct: 1 QPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQFAFRWFNCLLIREIPFNMVTRLWDT 180
Query: 371 YLAEGDALPDFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLPTQDWTHQELEMVLSRAF 430
YLAEGDALPDFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLPTQDWTHQELEMVLSRAF
Sbjct: 181 YLAEGDALPDFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLPTQDWTHQELEMVLSRAF 360
Query: 431 MWHSMFNNSPNHLAT 445
MWHSMFNNSPNHLAT
Sbjct: 361 MWHSMFNNSPNHLAT 405
>AV408566
Length = 423
Score = 225 bits (574), Expect = 1e-59
Identities = 112/112 (100%), Positives = 112/112 (100%)
Frame = +3
Query: 1 MKNNDEDNNNNPNIGIFDSRFSQTLRSVQGLLKGRSMPGKILLSQRVDPIDNSNLSSPTY 60
MKNNDEDNNNNPNIGIFDSRFSQTLRSVQGLLKGRSMPGKILLSQRVDPIDNSNLSSPTY
Sbjct: 87 MKNNDEDNNNNPNIGIFDSRFSQTLRSVQGLLKGRSMPGKILLSQRVDPIDNSNLSSPTY 266
Query: 61 TRSNSYNDAGTSDHASETVEVEVHSSSGIPGENKLKISTSHVENPSEDVRKS 112
TRSNSYNDAGTSDHASETVEVEVHSSSGIPGENKLKISTSHVENPSEDVRKS
Sbjct: 267 TRSNSYNDAGTSDHASETVEVEVHSSSGIPGENKLKISTSHVENPSEDVRKS 422
>TC15464
Length = 672
Score = 53.5 bits (127), Expect = 7e-08
Identities = 35/125 (28%), Positives = 62/125 (49%), Gaps = 5/125 (4%)
Frame = +2
Query: 300 LDGMQDHYTF----AQPGIQRLVFKLKELVRRIDDPVSTHMENQGLEFLQF-AFRWFNCL 354
L G +D++ + GI+ + +L +L++ D+ + H+E QF AFRW L
Sbjct: 2 LSGFRDNFVQQLDNSPVGIRATITRLSQLLKEHDEELWRHLEITSKVNPQFYAFRWITLL 181
Query: 355 LIREIPFNMVTRLWDTYLAEGDALPDFLVYIFASFLLTWSDKLQKLDFQDLVMFLQHLPT 414
L +E F +WDT L++ + + L+ I + L+ +L DF + LQ+ PT
Sbjct: 182 LTQEFNFADSIHIWDTILSDPEGPQETLLRICCAMLILIRKRLLAGDFTSNLKLLQNYPT 361
Query: 415 QDWTH 419
+ +H
Sbjct: 362 TNISH 376
>BP057230
Length = 541
Score = 45.4 bits (106), Expect = 2e-05
Identities = 23/57 (40%), Positives = 31/57 (54%), Gaps = 1/57 (1%)
Frame = -2
Query: 207 RQIAVDCPRTVPDV-SFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFFVV 262
R + D R P+ S+FQ Q L RIL W +RHP GY QG+++L+ P V
Sbjct: 360 RMVDQDLSRLYPEHGSYFQTPGCQGMLRRILLLWCLRHPECGYRQGMHELLAPLLYV 190
>TC19635
Length = 566
Score = 45.4 bits (106), Expect = 2e-05
Identities = 27/98 (27%), Positives = 50/98 (50%), Gaps = 1/98 (1%)
Frame = +3
Query: 323 ELVRRIDDPVSTHMENQGLEFLQF-AFRWFNCLLIREIPFNMVTRLWDTYLAEGDALPDF 381
+L++ D+ + H+E QF +FRW LL +E F+ + +WDT L++ + +
Sbjct: 3 QLLKEHDEELWRHLEVTTKVNPQFYSFRWITLLLTQEFNFHDILHIWDTLLSDPEGPQET 182
Query: 382 LVYIFASFLLTWSDKLQKLDFQDLVMFLQHLPTQDWTH 419
L+ I + L+ +L DF + LQ P+ + +H
Sbjct: 183 LLRICCAMLILVRRRLLAGDFTSNLKLLQSYPSTNISH 296
>BG662230
Length = 340
Score = 43.1 bits (100), Expect = 9e-05
Identities = 24/72 (33%), Positives = 40/72 (55%), Gaps = 5/72 (6%)
Frame = +3
Query: 202 EINMLRQIAVDCPRTVPDVSFFQQQQV-----QKSLERILYAWAIRHPASGYVQGINDLV 256
+ ++ QI D RT PD+ FF Q++L+ IL +A +P YVQG+N+++
Sbjct: 120 DTEIIDQIDRDVMRTHPDMHFFSGDSQFAKLNQEALKNILIIFAKLNPGVRYVQGMNEVL 299
Query: 257 TPFFVVFLSEYL 268
P F VF ++ +
Sbjct: 300 APLFYVFKNDLM 335
>TC12768 similar to UP|Q9FH10 (Q9FH10) Serine/threonine protein kinase-like,
partial (18%)
Length = 597
Score = 29.6 bits (65), Expect = 1.0
Identities = 20/51 (39%), Positives = 26/51 (50%), Gaps = 4/51 (7%)
Frame = +2
Query: 46 RVDPIDNSNLSSPTYTRSNSYNDAGTSDHASETVEVEV----HSSSGIPGE 92
R+ D SN S T+T S + AGT+ HA E++V SSS GE
Sbjct: 311 RISVTDLSNPGSTTFTEDLSISLAGTNIHAFTVAELKVITQHFSSSNFLGE 463
>BP052632
Length = 489
Score = 27.7 bits (60), Expect = 4.0
Identities = 39/146 (26%), Positives = 58/146 (39%), Gaps = 9/146 (6%)
Frame = -3
Query: 9 NNNPNIGIFDSRFSQTLRSVQGLLKGRSMPGK--ILLSQRVDPIDNS-NLS------SPT 59
N N NIG FD++F + +L G S K I+ + R ++ S N+ T
Sbjct: 406 NTNDNIGKFDNKFDKR------ILLGYSSNSKAYIVFNSRTQVVEESINVKFDD*SIEAT 245
Query: 60 YTRSNSYNDAGTSDHASETVEVEVHSSSGIPGENKLKISTSHVENPSEDVRKSSMGARAT 119
Y + + SD + EV ++ G P E TSH E PS + ++
Sbjct: 244 YRLEKDFLNLDLSDQ-----DEEVPTNEGQPSE------TSH-EEPSTQPQTTNPW---- 113
Query: 120 DSARVMKFTKVLSGTVVILDKLRELA 145
TK + +V LDKL A
Sbjct: 112 -----QGLTKQIHSLIVHLDKLTHFA 50
>TC9497
Length = 859
Score = 27.7 bits (60), Expect = 4.0
Identities = 23/91 (25%), Positives = 37/91 (40%)
Frame = +3
Query: 70 GTSDHASETVEVEVHSSSGIPGENKLKISTSHVENPSEDVRKSSMGARATDSARVMKFTK 129
G+S + + V +P +L ++ S P ++ D VMKF +
Sbjct: 144 GSSGNLVDVARQWVEKQGLLPSSEELPLTESF---PGSGGLIAAAKEARYDKEGVMKFFR 314
Query: 130 VLSGTVVILDKLRELAWSGVPDYMRPTVWRL 160
+L D+L E AWS +P RP R+
Sbjct: 315 MLIRRRYFSDELPEPAWS-IPSVQRPAQQRI 404
>AW428826
Length = 466
Score = 27.3 bits (59), Expect = 5.2
Identities = 13/36 (36%), Positives = 21/36 (58%)
Frame = +3
Query: 123 RVMKFTKVLSGTVVILDKLRELAWSGVPDYMRPTVW 158
R ++ T L TVVI+D++ +L P M+ +VW
Sbjct: 237 RCVERTSGLQRTVVIVDEIEDLKRGSEPRAMKASVW 344
>AV408165
Length = 424
Score = 27.3 bits (59), Expect = 5.2
Identities = 21/59 (35%), Positives = 31/59 (51%)
Frame = -1
Query: 103 ENPSEDVRKSSMGARATDSARVMKFTKVLSGTVVILDKLRELAWSGVPDYMRPTVWRLL 161
ENPS+ VRKS+ A + VM +T +G + LR +G P +M T WR++
Sbjct: 412 ENPSDSVRKST--AAGGGAPAVMIWTGFSNGLEAFVSMLRT---TGAP-HM*VTPWRVM 254
>BP043726
Length = 553
Score = 27.3 bits (59), Expect = 5.2
Identities = 14/24 (58%), Positives = 15/24 (62%), Gaps = 1/24 (4%)
Frame = +3
Query: 335 HMENQGLEFLQFAFRWFN-CLLIR 357
HMENQG E F WFN CL +R
Sbjct: 360 HMENQGKERQGRLFIWFNWCLPVR 431
>AV772949
Length = 521
Score = 26.9 bits (58), Expect = 6.8
Identities = 12/29 (41%), Positives = 15/29 (51%)
Frame = +1
Query: 155 PTVWRLLLGYAPPNSDRREGVLRRKRLEY 183
P L LGY PP+ R L +KR +Y
Sbjct: 364 PACGSLWLGYLPPDRGLRSSELAKKRTQY 450
>TC16838 similar to UP|BAD07764 (BAD07764) GTPase activating protein-like,
partial (16%)
Length = 586
Score = 26.9 bits (58), Expect = 6.8
Identities = 16/69 (23%), Positives = 31/69 (44%), Gaps = 2/69 (2%)
Frame = +1
Query: 346 FAFRWFNCLLIREIPFNMVTRLWDTYLAEGDALPDFLVYIFASFLLTWSDKL--QKLDFQ 403
F FRW RE + RLW+ + +Y+ + L + +++ +++DF
Sbjct: 22 FCFRWVLIPFKREFEYEDTMRLWEVLWTHYPS-EHLHLYVCVAVLKRYRNQIIGEEMDFD 198
Query: 404 DLVMFLQHL 412
L+ F+ L
Sbjct: 199 TLLKFINEL 225
>TC13685 similar to UP|Q9FJH5 (Q9FJH5) Gb|AAC98457.1 (At5g60800), partial
(37%)
Length = 585
Score = 26.9 bits (58), Expect = 6.8
Identities = 25/120 (20%), Positives = 50/120 (40%), Gaps = 2/120 (1%)
Frame = +2
Query: 19 SRFSQTLRSVQGL--LKGRSMPGKILLSQRVDPIDNSNLSSPTYTRSNSYNDAGTSDHAS 76
S+ + LR+ +G+ +K S GK+ +S +VDP D +
Sbjct: 227 SKIVKNLRAFKGVETVKAESNTGKVTVSGKVDP-------------------TKLRDSLA 349
Query: 77 ETVEVEVHSSSGIPGENKLKISTSHVENPSEDVRKSSMGARATDSARVMKFTKVLSGTVV 136
E ++ +V S P + K K EN +D ++ + T+ ++ K + ++ V+
Sbjct: 350 EKIKKKVELVSPQPKKEKEK-----AENKDKDTETNNKAEKKTEEKKINKDKQAVTTAVL 514
>BP055837
Length = 528
Score = 26.9 bits (58), Expect = 6.8
Identities = 28/94 (29%), Positives = 41/94 (42%)
Frame = -1
Query: 200 EDEINMLRQIAVDCPRTVPDVSFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPF 259
E+E+ L QIAV C VPD + V K +E + P V +++ +
Sbjct: 291 EEEMVALLQIAVACTAAVPD-QRPRMSHVVKMIEELRGVQV--SPCHDTVDSVSESPS-- 127
Query: 260 FVVFLSEYLEGSIDNWSMSDLSSDEISNVEADCY 293
LSE G N S+S L +D S ++ CY
Sbjct: 126 ----LSEDACGGGTN*SVSKLGNDLCSACKSICY 37
>AV425920
Length = 386
Score = 26.6 bits (57), Expect = 8.9
Identities = 13/40 (32%), Positives = 18/40 (44%)
Frame = +2
Query: 13 NIGIFDSRFSQTLRSVQGLLKGRSMPGKILLSQRVDPIDN 52
N D + Q L G GK+L ++R +PIDN
Sbjct: 257 NCAFVDFKNDMLASQAQRQLNGLRFLGKVLSAERANPIDN 376
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,672,081
Number of Sequences: 28460
Number of extensions: 100009
Number of successful extensions: 533
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 495
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 529
length of query: 445
length of database: 4,897,600
effective HSP length: 93
effective length of query: 352
effective length of database: 2,250,820
effective search space: 792288640
effective search space used: 792288640
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0106.13


