

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0105.15
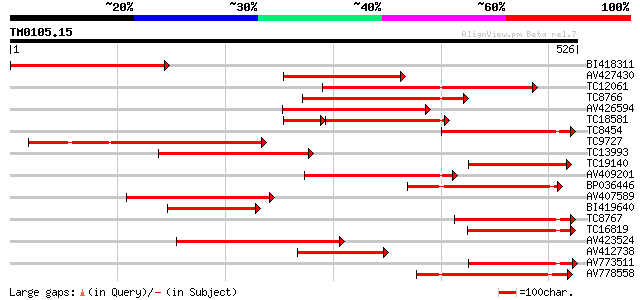
(526 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BI418311 297 3e-81
AV427430 228 2e-60
TC12061 similar to UP|Q9LN49 (Q9LN49) F18O14.21, partial (35%) 223 6e-59
TC8766 similar to UP|Q8LAZ1 (Q8LAZ1) Very-long-chain fatty acid ... 216 6e-57
AV426594 213 8e-56
TC18581 similar to UP|Q9FG87 (Q9FG87) Beta-ketoacyl-CoA synthase... 163 5e-55
TC8454 similar to UP|Q84UX3 (Q84UX3) Fatty acid elongase (Fragme... 204 4e-53
TC9727 weakly similar to UP|Q9FV67 (Q9FV67) Fatty acid elongase ... 203 6e-53
TC13993 similar to UP|Q8VWP9 (Q8VWP9) Fiddlehead-like protein, p... 202 1e-52
TC19140 similar to PIR|F86141|F86141 protein T25K16.11 [imported... 201 2e-52
AV409201 187 3e-48
BP036446 177 3e-45
AV407589 174 2e-44
BI419640 169 8e-43
TC8767 homologue to UP|Q8LAZ1 (Q8LAZ1) Very-long-chain fatty aci... 167 3e-42
TC16819 similar to UP|Q8LAZ1 (Q8LAZ1) Very-long-chain fatty acid... 149 1e-36
AV423524 147 5e-36
AV412738 144 5e-35
AV773511 144 5e-35
AV778558 142 1e-34
>BI418311
Length = 540
Score = 297 bits (760), Expect = 3e-81
Identities = 148/148 (100%), Positives = 148/148 (100%)
Frame = +1
Query: 1 MDSIDMDKERLTAEMDFKDSTSAVIKIRRRLPDFLQTVKLKYVKLGYGYTCNAATVLIFT 60
MDSIDMDKERLTAEMDFKDSTSAVIKIRRRLPDFLQTVKLKYVKLGYGYTCNAATVLIFT
Sbjct: 97 MDSIDMDKERLTAEMDFKDSTSAVIKIRRRLPDFLQTVKLKYVKLGYGYTCNAATVLIFT 276
Query: 61 FIFPLILFLLLQLTDLKLIHLSELWSDHTVHLDTDTVTGSAVLLFLFGLYWAKRSAPVYL 120
FIFPLILFLLLQLTDLKLIHLSELWSDHTVHLDTDTVTGSAVLLFLFGLYWAKRSAPVYL
Sbjct: 277 FIFPLILFLLLQLTDLKLIHLSELWSDHTVHLDTDTVTGSAVLLFLFGLYWAKRSAPVYL 456
Query: 121 VDFACYKPEKERKISVDSFLKMTEETGG 148
VDFACYKPEKERKISVDSFLKMTEETGG
Sbjct: 457 VDFACYKPEKERKISVDSFLKMTEETGG 540
>AV427430
Length = 345
Score = 228 bits (581), Expect = 2e-60
Identities = 112/113 (99%), Positives = 112/113 (99%)
Frame = +2
Query: 255 SAGLISIDLAKDLLKANPNSYAVVVSTENITLNWYFGNDRSMLLCNCIFRMGGAAVLLSN 314
SAGLISIDLAKDLLKANPNSYAVVVSTENITLNWYFGNDRSMLLCNCIFRMGGAAVLLSN
Sbjct: 5 SAGLISIDLAKDLLKANPNSYAVVVSTENITLNWYFGNDRSMLLCNCIFRMGGAAVLLSN 184
Query: 315 KSSDRAKSKYELVHTVRTHKGSDDKHYNCVYQMEDEKGKVGVCLARELMAVAG 367
KSSDRAKSKYELVHTVRTHKG DDKHYNCVYQMEDEKGKVGVCLARELMAVAG
Sbjct: 185 KSSDRAKSKYELVHTVRTHKGFDDKHYNCVYQMEDEKGKVGVCLARELMAVAG 343
>TC12061 similar to UP|Q9LN49 (Q9LN49) F18O14.21, partial (35%)
Length = 613
Score = 223 bits (568), Expect = 6e-59
Identities = 111/200 (55%), Positives = 148/200 (73%), Gaps = 1/200 (0%)
Frame = +1
Query: 291 GNDRSMLLCNCIFRMGGAAVLLSNKSSDRAKSKYELVHTVRTHKGSDDKHYNCVYQMEDE 350
GN +SML+ NC+FR+GG+AVLLSNK+SD+ ++KY+LVH VRTHKG+DDK + CVYQ +DE
Sbjct: 7 GNKKSMLIPNCLFRVGGSAVLLSNKASDKFRAKYKLVHVVRTHKGADDKAFRCVYQEQDE 186
Query: 351 KGKVGVCLARELMAVAGDALKTNITTLGPLVLPFMEQFMFFVSLVRRKMTFLKRSKPYIP 410
GK GV L+++LMA+AG ALKTNITTLGPLVLP EQ +FF +L+ +K+ F ++KPYIP
Sbjct: 187 AGKTGVSLSKDLMAIAGGALKTNITTLGPLVLPVSEQLLFFATLIVKKV-FNAKAKPYIP 363
Query: 411 DFKLAFEHFCIHAGGRAVLDEMQKNLELSEWHMEPSRM-TLHRFGNTSSSSLWYELAYTE 469
DFKLAF+HFCIHAGGRAV+DE++KNL+L H+E S+ T +G + +
Sbjct: 364 DFKLAFDHFCIHAGGRAVIDELEKNLQLLPGHVEASKNDTA*VWGTLLQAQFGMNWVTLK 543
Query: 470 AKGRVGKGDRVWQIAFGSGF 489
KG+ + FG+GF
Sbjct: 544 PKGKDXEXKXNLAXCFGNGF 603
>TC8766 similar to UP|Q8LAZ1 (Q8LAZ1) Very-long-chain fatty acid condensing
enzyme CUT1, partial (31%)
Length = 462
Score = 216 bits (551), Expect = 6e-57
Identities = 103/154 (66%), Positives = 131/154 (84%)
Frame = +3
Query: 272 PNSYAVVVSTENITLNWYFGNDRSMLLCNCIFRMGGAAVLLSNKSSDRAKSKYELVHTVR 331
PNS AVVVSTE IT N+Y GN+R+MLL NC+FRMGGAA++LSN+SS+ +++KY+LVH VR
Sbjct: 3 PNSNAVVVSTEIITPNYYQGNERAMLLPNCLFRMGGAAIMLSNRSSESSRAKYKLVHVVR 182
Query: 332 THKGSDDKHYNCVYQMEDEKGKVGVCLARELMAVAGDALKTNITTLGPLVLPFMEQFMFF 391
THKGSDDK Y CV++ ED++GKVG+ L ++LMA+AG+ALK+NITT+GPLVLP EQ +F
Sbjct: 183 THKGSDDKAYRCVFEEEDKEGKVGISLQKDLMAIAGEALKSNITTMGPLVLPASEQVLFL 362
Query: 392 VSLVRRKMTFLKRSKPYIPDFKLAFEHFCIHAGG 425
+L+ RK+ F + KPYIPDFK AFEHFCIHAGG
Sbjct: 363 ATLIGRKI-FNPKWKPYIPDFKQAFEHFCIHAGG 461
>AV426594
Length = 415
Score = 213 bits (541), Expect = 8e-56
Identities = 104/137 (75%), Positives = 120/137 (86%)
Frame = +1
Query: 254 CSAGLISIDLAKDLLKANPNSYAVVVSTENITLNWYFGNDRSMLLCNCIFRMGGAAVLLS 313
CSAG+ISIDLAK+LL+ +PNSYA+VVS ENITLNWY GNDRS L+ NC+FRMGGAA+LLS
Sbjct: 4 CSAGIISIDLAKELLQVHPNSYALVVSMENITLNWYPGNDRSKLVSNCLFRMGGAAILLS 183
Query: 314 NKSSDRAKSKYELVHTVRTHKGSDDKHYNCVYQMEDEKGKVGVCLARELMAVAGDALKTN 373
NKSSDR +SKY+LV TVRTHKGSDDK ++CV Q ED GK GV L+++LMAVAGDALKTN
Sbjct: 184 NKSSDRRRSKYQLVDTVRTHKGSDDKCFSCVTQEEDANGKTGVTLSKDLMAVAGDALKTN 363
Query: 374 ITTLGPLVLPFMEQFMF 390
ITTLGPLVLP EQ +F
Sbjct: 364 ITTLGPLVLPTSEQLLF 414
>TC18581 similar to UP|Q9FG87 (Q9FG87) Beta-ketoacyl-CoA synthase
(AT5g43760/MQD19_11), partial (28%)
Length = 465
Score = 163 bits (413), Expect(2) = 5e-55
Identities = 80/115 (69%), Positives = 97/115 (83%)
Frame = +2
Query: 294 RSMLLCNCIFRMGGAAVLLSNKSSDRAKSKYELVHTVRTHKGSDDKHYNCVYQMEDEKGK 353
R ML+ NC+FRMGGAA+LLS+KSSDR ++KY+LVHTVRTHKG+DDK Y CV+Q EDE
Sbjct: 122 RPMLVSNCLFRMGGAAILLSSKSSDRRRAKYQLVHTVRTHKGADDKSYGCVFQEEDETKT 301
Query: 354 VGVCLARELMAVAGDALKTNITTLGPLVLPFMEQFMFFVSLVRRKMTFLKRSKPY 408
VGV L+++LMAVAG+ALKTNITTLGPLVLP EQ +FFV+LV RK+ F + KPY
Sbjct: 302 VGVALSKDLMAVAGEALKTNITTLGPLVLPMSEQILFFVTLVARKV-FKMKIKPY 463
Score = 68.6 bits (166), Expect(2) = 5e-55
Identities = 32/39 (82%), Positives = 36/39 (92%)
Frame = +1
Query: 255 SAGLISIDLAKDLLKANPNSYAVVVSTENITLNWYFGND 293
SAGLISIDLAK LL+ +PNSYA+VVS ENITLNWYFGN+
Sbjct: 4 SAGLISIDLAKQLLQVHPNSYALVVSMENITLNWYFGNN 120
>TC8454 similar to UP|Q84UX3 (Q84UX3) Fatty acid elongase (Fragment),
partial (50%)
Length = 661
Score = 204 bits (518), Expect = 4e-53
Identities = 94/125 (75%), Positives = 109/125 (87%)
Frame = +1
Query: 401 FLKRSKPYIPDFKLAFEHFCIHAGGRAVLDEMQKNLELSEWHMEPSRMTLHRFGNTSSSS 460
F + KPYIPDFKLAFEHFCIHAGGRAVLDE++KNL+LS+WHMEPSRMTL+RFGNTSSSS
Sbjct: 7 FKVKIKPYIPDFKLAFEHFCIHAGGRAVLDELEKNLDLSDWHMEPSRMTLNRFGNTSSSS 186
Query: 461 LWYELAYTEAKGRVGKGDRVWQIAFGSGFKCNSAVWKAVRDMPELGDWRGNPWDDSIDKY 520
LWYELAYTEAKGR+ KGDR WQIAFGSGFKCNSAVW+A++ + + NPW D I ++
Sbjct: 187 LWYELAYTEAKGRIRKGDRTWQIAFGSGFKCNSAVWQALKTINPAKE--KNPWMDEIHEF 360
Query: 521 PVNVP 525
PV+VP
Sbjct: 361 PVHVP 375
>TC9727 weakly similar to UP|Q9FV67 (Q9FV67) Fatty acid elongase 1-like
protein, partial (40%)
Length = 731
Score = 203 bits (516), Expect = 6e-53
Identities = 107/221 (48%), Positives = 142/221 (63%)
Frame = +2
Query: 18 KDSTSAVIKIRRRLPDFLQTVKLKYVKLGYGYTCNAATVLIFTFIFPLILFLLLQLTDLK 77
K T + R LPDF +VKLKYVKLGY Y ++ F+ PL++ + QL+
Sbjct: 80 KPDTPLMPPSSRNLPDFKNSVKLKYVKLGYHYLITHG---MYLFLSPLVVLIAAQLSTFS 250
Query: 78 LIHLSELWSDHTVHLDTDTVTGSAVLLFLFGLYWAKRSAPVYLVDFACYKPEKERKISVD 137
+ + ++W + +L + S +L+FL LY+ R PVYLV+F+CYKPE+ RK +
Sbjct: 251 IKDIYDIWDNLQYNL-ISVIICSTLLVFLSTLYFLTRPKPVYLVNFSCYKPEEARKCTKK 427
Query: 138 SFLKMTEETGGFEEETLQFQRRISMKAGLGDETYLPRGITSRPPNLCMKEARLEAEAVMF 197
F+ + +G F EE L FQR+I ++GLG+ TY P + + PPN M EAR EAEAVM+
Sbjct: 428 KFMDHSRMSGFFTEENLDFQRKILERSGLGENTYFPEAVLNIPPNPTMHEARKEAEAVMY 607
Query: 198 GAMDALFAKTGVNPKDIDILVVNCSLFNPTPSLSAMIVNHY 238
GA+D LFAKT V KDI IL+VNCSLF PTPSLSAMIVNHY
Sbjct: 608 GAIDELFAKTSVKAKDIGILIVNCSLFCPTPSLSAMIVNHY 730
>TC13993 similar to UP|Q8VWP9 (Q8VWP9) Fiddlehead-like protein, partial
(26%)
Length = 437
Score = 202 bits (514), Expect = 1e-52
Identities = 94/145 (64%), Positives = 120/145 (81%), Gaps = 1/145 (0%)
Frame = +3
Query: 139 FLKMTEETGGFEEETLQFQRRISMKAGLGDETYLPRGITSRPPNLC-MKEARLEAEAVMF 197
++++ ++G F+EE+L FQ+RI M +G+GDETY+P+ + S N MKE R EA VMF
Sbjct: 3 YIELASKSGKFDEESLAFQKRILMSSGIGDETYIPKAVVSSTTNTATMKEGRAEASTVMF 182
Query: 198 GAMDALFAKTGVNPKDIDILVVNCSLFNPTPSLSAMIVNHYKLRTNIKSYNLGGMGCSAG 257
GA+D LF KTG+ PKDI ILVVNCS+FNPTPSLSAMI+NHYK+R NI SYNLGGMGCSAG
Sbjct: 183 GALDELFEKTGIRPKDIGILVVNCSIFNPTPSLSAMIINHYKMRGNILSYNLGGMGCSAG 362
Query: 258 LISIDLAKDLLKANPNSYAVVVSTE 282
+I +DLA+D+L++NPN+YAVVVSTE
Sbjct: 363 IIGVDLARDILQSNPNNYAVVVSTE 437
>TC19140 similar to PIR|F86141|F86141 protein T25K16.11 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(14%)
Length = 525
Score = 201 bits (512), Expect = 2e-52
Identities = 94/96 (97%), Positives = 94/96 (97%)
Frame = +2
Query: 426 RAVLDEMQKNLELSEWHMEPSRMTLHRFGNTSSSSLWYELAYTEAKGRVGKGDRVWQIAF 485
RAVLDEMQKNLELSEWHMEPSRMTLHRFGNTSSSSLWYELAYTEAK RVGKGDRV QIAF
Sbjct: 2 RAVLDEMQKNLELSEWHMEPSRMTLHRFGNTSSSSLWYELAYTEAKARVGKGDRV*QIAF 181
Query: 486 GSGFKCNSAVWKAVRDMPELGDWRGNPWDDSIDKYP 521
GSGFKCNSAVWKAVRDMPELGDWRGNPWDDSIDKYP
Sbjct: 182 GSGFKCNSAVWKAVRDMPELGDWRGNPWDDSIDKYP 289
>AV409201
Length = 426
Score = 187 bits (476), Expect = 3e-48
Identities = 91/142 (64%), Positives = 118/142 (83%)
Frame = +2
Query: 274 SYAVVVSTENITLNWYFGNDRSMLLCNCIFRMGGAAVLLSNKSSDRAKSKYELVHTVRTH 333
S AVVVSTE IT N+Y GN+R+MLL NC+FRMGGAA+LL+N+ S + ++KY LVH VRTH
Sbjct: 2 SNAVVVSTEIITPNYYQGNERAMLLPNCLFRMGGAAILLTNRKSAKTRAKYRLVHVVRTH 181
Query: 334 KGSDDKHYNCVYQMEDEKGKVGVCLARELMAVAGDALKTNITTLGPLVLPFMEQFMFFVS 393
KGSDDK Y CVY+ ED++GKVG+ L+++LMA+AG+ALK+NITT+GP+VLP EQ +F V+
Sbjct: 182 KGSDDKAYRCVYEEEDKEGKVGISLSKDLMAIAGEALKSNITTIGPIVLPASEQLIFLVT 361
Query: 394 LVRRKMTFLKRSKPYIPDFKLA 415
L+ RK+ F + KPYIPDFK A
Sbjct: 362 LIGRKI-FNPKWKPYIPDFKQA 424
>BP036446
Length = 522
Score = 177 bits (450), Expect = 3e-45
Identities = 93/144 (64%), Positives = 109/144 (75%)
Frame = -1
Query: 370 LKTNITTLGPLVLPFMEQFMFFVSLVRRKMTFLKRSKPYIPDFKLAFEHFCIHAGGRAVL 429
LK NITTLGPLVLP EQF FF +LV R+ K++KPYIPD+KLAFEH C+ A + VL
Sbjct: 522 LKDNITTLGPLVLPVSEQFHFFTNLVFRR----KKTKPYIPDYKLAFEHVCVVATSKKVL 355
Query: 430 DEMQKNLELSEWHMEPSRMTLHRFGNTSSSSLWYELAYTEAKGRVGKGDRVWQIAFGSGF 489
DE+QKNLEL+E +ME SR TL RFGNTSSSS WYELAY E + RV KGDRV QIA GSGF
Sbjct: 354 DEVQKNLELTEEYMEASRKTLERFGNTSSSSNWYELAYLEFEERVQKGDRVCQIAAGSGF 175
Query: 490 KCNSAVWKAVRDMPELGDWRGNPW 513
CNS VWKA+++ +G +PW
Sbjct: 174 MCNSVVWKALKN---VGRPNQSPW 112
>AV407589
Length = 418
Score = 174 bits (442), Expect = 2e-44
Identities = 88/137 (64%), Positives = 103/137 (74%)
Frame = +1
Query: 109 LYWAKRSAPVYLVDFACYKPEKERKISVDSFLKMTEETGGFEEETLQFQRRISMKAGLGD 168
LY+ R VYLVDF+CYKPE E ++ + F++ +E TG F EE L FQ++I ++GLG
Sbjct: 7 LYFMSRPRGVYLVDFSCYKPEPECNVTREIFMQRSELTGTFTEENLAFQKKILERSGLGQ 186
Query: 169 ETYLPRGITSRPPNLCMKEARLEAEAVMFGAMDALFAKTGVNPKDIDILVVNCSLFNPTP 228
+TYLP I S PPN CM EAR EAE VMFGA+D L KTGV KDI ILVVNCSLFNPTP
Sbjct: 187 KTYLPPAILSIPPNPCMAEARKEAEQVMFGAIDQLLEKTGVKAKDIGILVVNCSLFNPTP 366
Query: 229 SLSAMIVNHYKLRTNIK 245
SLSAMIVNHYKLR NI+
Sbjct: 367 SLSAMIVNHYKLRGNIQ 417
>BI419640
Length = 264
Score = 169 bits (429), Expect = 8e-43
Identities = 84/86 (97%), Positives = 84/86 (97%)
Frame = +3
Query: 147 GGFEEETLQFQRRISMKAGLGDETYLPRGITSRPPNLCMKEARLEAEAVMFGAMDALFAK 206
GGF EETL FQRRISMKAGLGDETYLPRGITSRPPNLCMKEARLEAEAVMFGAMDALFAK
Sbjct: 3 GGFGEETLPFQRRISMKAGLGDETYLPRGITSRPPNLCMKEARLEAEAVMFGAMDALFAK 182
Query: 207 TGVNPKDIDILVVNCSLFNPTPSLSA 232
TGVNPKDIDILVVNCSLFNPTPSLSA
Sbjct: 183 TGVNPKDIDILVVNCSLFNPTPSLSA 260
>TC8767 homologue to UP|Q8LAZ1 (Q8LAZ1) Very-long-chain fatty acid
condensing enzyme CUT1, partial (23%)
Length = 770
Score = 167 bits (424), Expect = 3e-42
Identities = 80/113 (70%), Positives = 92/113 (80%)
Frame = +1
Query: 413 KLAFEHFCIHAGGRAVLDEMQKNLELSEWHMEPSRMTLHRFGNTSSSSLWYELAYTEAKG 472
K AFEHFCIHAGGRAV+DE+Q+NL+LS H+E SRMTLHRFGNTSSSSLWYEL Y E+KG
Sbjct: 1 KQAFEHFCIHAGGRAVIDELQRNLQLSAEHVEASRMTLHRFGNTSSSSLWYELNYIESKG 180
Query: 473 RVGKGDRVWQIAFGSGFKCNSAVWKAVRDMPELGDWRGNPWDDSIDKYPVNVP 525
R+ +GDRVWQIAFGSGFKCNSAVWK R + D P D ID+YPV++P
Sbjct: 181 RMKRGDRVWQIAFGSGFKCNSAVWKCNRTIKTPFD---GPCSDCIDRYPVHIP 330
>TC16819 similar to UP|Q8LAZ1 (Q8LAZ1) Very-long-chain fatty acid condensing
enzyme CUT1, partial (21%)
Length = 723
Score = 149 bits (375), Expect = 1e-36
Identities = 69/101 (68%), Positives = 82/101 (80%)
Frame = +1
Query: 425 GRAVLDEMQKNLELSEWHMEPSRMTLHRFGNTSSSSLWYELAYTEAKGRVGKGDRVWQIA 484
GRAV+DE+QKNL+LS H+E SRMTLHRFGNTSSSSLWYEL Y E+KGR+ +GDR+WQI
Sbjct: 1 GRAVIDELQKNLQLSAEHVEASRMTLHRFGNTSSSSLWYELNYIESKGRMKRGDRIWQIG 180
Query: 485 FGSGFKCNSAVWKAVRDMPELGDWRGNPWDDSIDKYPVNVP 525
FGSGFKCNSAVWK R + D PW+D ID+YPV++P
Sbjct: 181 FGSGFKCNSAVWKCNRSIKTPVD---GPWEDCIDRYPVHIP 294
>AV423524
Length = 528
Score = 147 bits (370), Expect = 5e-36
Identities = 69/157 (43%), Positives = 107/157 (67%), Gaps = 1/157 (0%)
Frame = +2
Query: 155 QFQRRISMKAGLGDETYLPRGI-TSRPPNLCMKEARLEAEAVMFGAMDALFAKTGVNPKD 213
+F + + +G+G++TY PR + R + +++ LE E ++ L +T ++P +
Sbjct: 56 KFLLKAIVSSGIGEQTYAPRNVFDGREASPTLEDGILEMEEFFNDSIAKLLQRTSISPSE 235
Query: 214 IDILVVNCSLFNPTPSLSAMIVNHYKLRTNIKSYNLGGMGCSAGLISIDLAKDLLKANPN 273
ID+LVVN S+F PSLS+ I+N YKLR +IK YNL GMGCSA LIS+D+ +++ K N
Sbjct: 236 IDVLVVNISMFAAVPSLSSRIINRYKLRHDIKVYNLTGMGCSASLISVDIVQNIFKTQKN 415
Query: 274 SYAVVVSTENITLNWYFGNDRSMLLCNCIFRMGGAAV 310
A++V++E+++ NWY GNDRSM+L NC+FR GG A+
Sbjct: 416 KLALLVTSESLSPNWYNGNDRSMILANCLFRSGGCAM 526
>AV412738
Length = 397
Score = 144 bits (362), Expect = 5e-35
Identities = 64/84 (76%), Positives = 77/84 (91%)
Frame = +3
Query: 268 LKANPNSYAVVVSTENITLNWYFGNDRSMLLCNCIFRMGGAAVLLSNKSSDRAKSKYELV 327
++ +PNSYA+VVS ENITLNWYFGNDRSML+ NC+FRMGGAAVLLSNKSSDR+++KY+L+
Sbjct: 135 MQVHPNSYALVVSMENITLNWYFGNDRSMLVSNCLFRMGGAAVLLSNKSSDRSRAKYQLI 314
Query: 328 HTVRTHKGSDDKHYNCVYQMEDEK 351
HTVRTHKG+DDK Y CV+Q ED K
Sbjct: 315 HTVRTHKGADDKSYGCVFQEEDAK 386
>AV773511
Length = 463
Score = 144 bits (362), Expect = 5e-35
Identities = 68/101 (67%), Positives = 83/101 (81%)
Frame = -2
Query: 426 RAVLDEMQKNLELSEWHMEPSRMTLHRFGNTSSSSLWYELAYTEAKGRVGKGDRVWQIAF 485
RAV+DE++KNL+L H+E SRMTLHRFG TSSSS+WYELAY EAKGRV KG+R+WQIAF
Sbjct: 462 RAVIDELEKNLQLLPVHVEASRMTLHRFGYTSSSSIWYELAYIEAKGRVRKGNRLWQIAF 283
Query: 486 GSGFKCNSAVWKAVRDMPELGDWRGNPWDDSIDKYPVNVPS 526
GSGFKCNSAVW+A+R++ + PW+D IDKYPV + S
Sbjct: 282 GSGFKCNSAVWEALRNVRSSPN---GPWEDCIDKYPVEIDS 169
>AV778558
Length = 444
Score = 142 bits (359), Expect = 1e-34
Identities = 72/145 (49%), Positives = 98/145 (66%)
Frame = -2
Query: 378 GPLVLPFMEQFMFFVSLVRRKMTFLKRSKPYIPDFKLAFEHFCIHAGGRAVLDEMQKNLE 437
G +LP E+F + VS++++K F+K Y+P+FK +HFC+ GRAV+ E+ K L+
Sbjct: 443 GSEILPPSEKFWYGVSVIQKK--FIKSEGIYVPNFKTVIQHFCLPCSGRAVIREVGKGLK 270
Query: 438 LSEWHMEPSRMTLHRFGNTSSSSLWYELAYTEAKGRVGKGDRVWQIAFGSGFKCNSAVWK 497
L+E +EP+ MTL+RFG+ SSSSLWYELAY EAK RV KGD+VW + GSG KC V K
Sbjct: 269 LAERDIEPAMMTLYRFGDPSSSSLWYELAYLEAKERVQKGDKVWPLGMGSGPKCIRVVLK 90
Query: 498 AVRDMPELGDWRGNPWDDSIDKYPV 522
VR P +G+ + PW D I YP+
Sbjct: 89 CVR--PLVGESQKGPWADCIHPYPI 21
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.137 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,626,428
Number of Sequences: 28460
Number of extensions: 139224
Number of successful extensions: 800
Number of sequences better than 10.0: 74
Number of HSP's better than 10.0 without gapping: 778
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 781
length of query: 526
length of database: 4,897,600
effective HSP length: 94
effective length of query: 432
effective length of database: 2,222,360
effective search space: 960059520
effective search space used: 960059520
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0105.15


