

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
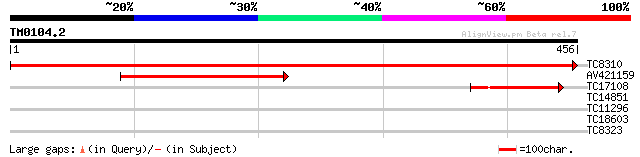
Query= TM0104.2
(456 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8310 UP|Q9SPM8 (Q9SPM8) Nod factor binding lectin-nucleotide p... 907 0.0
AV421159 174 3e-44
TC17108 homologue to UP|Q84UD8 (Q84UD8) Apyrase-like protein, pa... 103 6e-23
TC14851 similar to UP|Q9ZTX0 (Q9ZTX0) Similarity to SCAMP37, par... 27 7.0
TC11296 weakly similar to UP|Q9LMB9 (Q9LMB9) F14D16.24, partial ... 27 7.0
TC18603 similar to UP|Q9SJF1 (Q9SJF1) T27G7.9, partial (15%) 27 9.1
TC8323 weakly similar to UP|Q8R419 (Q8R419) Endoribonuclease Dic... 27 9.1
>TC8310 UP|Q9SPM8 (Q9SPM8) Nod factor binding lectin-nucleotide
phosphohydrolase, complete
Length = 1489
Score = 907 bits (2345), Expect = 0.0
Identities = 456/456 (100%), Positives = 456/456 (100%)
Frame = +1
Query: 1 MDFLISLMTFVFMLMPAISSSQYLGNNILMNRKILLPKNQEPVTSYAVIFDAGSTGSRVH 60
MDFLISLMTFVFMLMPAISSSQYLGNNILMNRKILLPKNQEPVTSYAVIFDAGSTGSRVH
Sbjct: 43 MDFLISLMTFVFMLMPAISSSQYLGNNILMNRKILLPKNQEPVTSYAVIFDAGSTGSRVH 222
Query: 61 VYNFDQNLDLLPVENELEFYDSVKPGLSSYAANPEEAAESLIPLLKEAENVVPVSQQPNT 120
VYNFDQNLDLLPVENELEFYDSVKPGLSSYAANPEEAAESLIPLLKEAENVVPVSQQPNT
Sbjct: 223 VYNFDQNLDLLPVENELEFYDSVKPGLSSYAANPEEAAESLIPLLKEAENVVPVSQQPNT 402
Query: 121 PVKLGATAGLRLLEGNAAENILQAVRDMLSNRSALNVQSDAVSILDGTQEGSYLWVTINY 180
PVKLGATAGLRLLEGNAAENILQAVRDMLSNRSALNVQSDAVSILDGTQEGSYLWVTINY
Sbjct: 403 PVKLGATAGLRLLEGNAAENILQAVRDMLSNRSALNVQSDAVSILDGTQEGSYLWVTINY 582
Query: 181 LLGKLGKRFTKTVGVVDLGGGSVQMTYAVSRNTAKNAPKVPEGEDPYIKKLVLQGKKYDL 240
LLGKLGKRFTKTVGVVDLGGGSVQMTYAVSRNTAKNAPKVPEGEDPYIKKLVLQGKKYDL
Sbjct: 583 LLGKLGKRFTKTVGVVDLGGGSVQMTYAVSRNTAKNAPKVPEGEDPYIKKLVLQGKKYDL 762
Query: 241 YVHSYLRYGREAFRAEIFKVAGGSANPCILAGFDGAYTYSGAEYKVSAPASGSNLNQCRK 300
YVHSYLRYGREAFRAEIFKVAGGSANPCILAGFDGAYTYSGAEYKVSAPASGSNLNQCRK
Sbjct: 763 YVHSYLRYGREAFRAEIFKVAGGSANPCILAGFDGAYTYSGAEYKVSAPASGSNLNQCRK 942
Query: 301 IALKALKVNAPCPYQNCTFGGIWNGGGGSGQKNLFLTSSFYYLSEDVGIFVNKPNAKIRP 360
IALKALKVNAPCPYQNCTFGGIWNGGGGSGQKNLFLTSSFYYLSEDVGIFVNKPNAKIRP
Sbjct: 943 IALKALKVNAPCPYQNCTFGGIWNGGGGSGQKNLFLTSSFYYLSEDVGIFVNKPNAKIRP 1122
Query: 361 VDLKTAAKLACKTNLEDAKSKYPDLYEKDSVEYVCLDLVYVYTLLVDGFGLDPFQEVTVA 420
VDLKTAAKLACKTNLEDAKSKYPDLYEKDSVEYVCLDLVYVYTLLVDGFGLDPFQEVTVA
Sbjct: 1123VDLKTAAKLACKTNLEDAKSKYPDLYEKDSVEYVCLDLVYVYTLLVDGFGLDPFQEVTVA 1302
Query: 421 NEIEYQDALVEAAWPLGTAIEAISSLPKFERLMYFI 456
NEIEYQDALVEAAWPLGTAIEAISSLPKFERLMYFI
Sbjct: 1303NEIEYQDALVEAAWPLGTAIEAISSLPKFERLMYFI 1410
>AV421159
Length = 410
Score = 174 bits (441), Expect = 3e-44
Identities = 85/135 (62%), Positives = 109/135 (79%)
Frame = +2
Query: 90 YAANPEEAAESLIPLLKEAENVVPVSQQPNTPVKLGATAGLRLLEGNAAENILQAVRDML 149
YA NP++AAESL+ LL +AE+VVP + T V++GATAGLR LEG+A++ ILQAVRD+L
Sbjct: 5 YAQNPQQAAESLVSLLDKAESVVPRELRSKTQVRVGATAGLRALEGDASDRILQAVRDLL 184
Query: 150 SNRSALNVQSDAVSILDGTQEGSYLWVTINYLLGKLGKRFTKTVGVVDLGGGSVQMTYAV 209
RS+L ++DAV++LDG QEG++ WVTINYLLG LGK ++KTVGVVDLGGGSVQM YA+
Sbjct: 185 KKRSSLKSEADAVTVLDGVQEGAFQWVTINYLLGNLGKDYSKTVGVVDLGGGSVQMAYAI 364
Query: 210 SRNTAKNAPKVPEGE 224
S A APKV +G+
Sbjct: 365 SETEAAQAPKVTDGQ 409
>TC17108 homologue to UP|Q84UD8 (Q84UD8) Apyrase-like protein, partial (16%)
Length = 573
Score = 103 bits (257), Expect = 6e-23
Identities = 48/75 (64%), Positives = 62/75 (82%)
Frame = +1
Query: 371 CKTNLEDAKSKYPDLYEKDSVEYVCLDLVYVYTLLVDGFGLDPFQEVTVANEIEYQDALV 430
C+T L DAKS YP + E ++ Y+C+DLVY YTLLVDGFG+ P+QEVT+ +++Y DALV
Sbjct: 16 CQTKLGDAKSTYPHI-EDGNLPYICMDLVYQYTLLVDGFGIYPWQEVTLVKKVKYDDALV 192
Query: 431 EAAWPLGTAIEAISS 445
EAAWPLG+AIEA+SS
Sbjct: 193 EAAWPLGSAIEAVSS 237
>TC14851 similar to UP|Q9ZTX0 (Q9ZTX0) Similarity to SCAMP37, partial (47%)
Length = 708
Score = 26.9 bits (58), Expect = 7.0
Identities = 9/16 (56%), Positives = 13/16 (81%)
Frame = +2
Query: 385 LYEKDSVEYVCLDLVY 400
LY+K +YVC+D+VY
Sbjct: 404 LYDKSPCDYVCIDVVY 451
>TC11296 weakly similar to UP|Q9LMB9 (Q9LMB9) F14D16.24, partial (7%)
Length = 493
Score = 26.9 bits (58), Expect = 7.0
Identities = 12/37 (32%), Positives = 19/37 (50%)
Frame = +1
Query: 347 VGIFVNKPNAKIRPVDLKTAAKLACKTNLEDAKSKYP 383
VG+ +PNAK+RP + KL +++D P
Sbjct: 88 VGLLCVQPNAKLRPRMSEVVEKLTVNVDMKDVSISKP 198
>TC18603 similar to UP|Q9SJF1 (Q9SJF1) T27G7.9, partial (15%)
Length = 714
Score = 26.6 bits (57), Expect = 9.1
Identities = 14/44 (31%), Positives = 22/44 (49%)
Frame = +3
Query: 264 SANPCILAGFDGAYTYSGAEYKVSAPASGSNLNQCRKIALKALK 307
SA PC+L + +Y+YS YK C +++ KAL+
Sbjct: 579 SAKPCLLDPYTHSYSYSEVNYK-----------*CLRVSCKALR 677
>TC8323 weakly similar to UP|Q8R419 (Q8R419) Endoribonuclease Dicer
(Fragment), partial (8%)
Length = 639
Score = 26.6 bits (57), Expect = 9.1
Identities = 18/41 (43%), Positives = 20/41 (47%)
Frame = +3
Query: 360 PVDLKTAAKLACKTNLEDAKSKYPDLYEKDSVEYVCLDLVY 400
P D KTA KLA K E KS Y + V + CL VY
Sbjct: 375 PADRKTAQKLASK---EILKSLKQ*TYLRTQVMFTCLVCVY 488
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.317 0.136 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,785,497
Number of Sequences: 28460
Number of extensions: 83411
Number of successful extensions: 423
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 420
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 422
length of query: 456
length of database: 4,897,600
effective HSP length: 93
effective length of query: 363
effective length of database: 2,250,820
effective search space: 817047660
effective search space used: 817047660
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0104.2


