

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
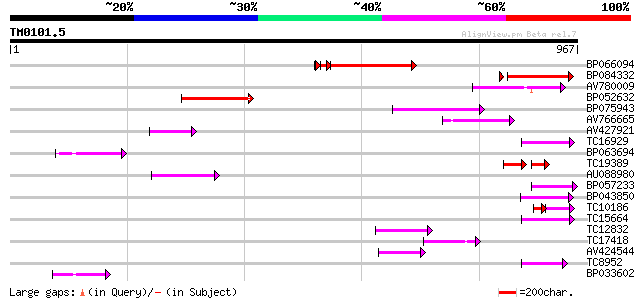
Query= TM0101.5
(967 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BP066094 270 2e-77
BP084332 183 1e-46
AV780009 117 7e-27
BP052632 107 9e-24
BP075943 65 5e-11
AV766665 64 9e-11
AV427921 64 9e-11
TC16929 weakly similar to UP|Q9LFY6 (Q9LFY6) T7N9.5, partial (4%) 63 3e-10
BP063694 62 3e-10
TC19389 weakly similar to UP|POLX_TOBAC (P10978) Retrovirus-rela... 42 1e-09
AU088980 60 2e-09
BP057233 60 2e-09
BP043850 57 2e-08
TC10186 weakly similar to UP|Q9FH39 (Q9FH39) Copia-type polyprot... 39 4e-07
TC15664 weakly similar to UP|O81617 (O81617) F8M12.17 protein, p... 52 6e-07
TC12832 weakly similar to UP|POLX_TOBAC (P10978) Retrovirus-rela... 50 1e-06
TC17418 similar to UP|Q94AX2 (Q94AX2) AT5g39790/MKM21_80, partia... 47 1e-05
AV424544 46 3e-05
TC8952 similar to UP|O82607 (O82607) T2L5.9 protein, partial (3%) 45 4e-05
BP033602 44 2e-04
>BP066094
Length = 532
Score = 270 bits (691), Expect(3) = 2e-77
Identities = 135/149 (90%), Positives = 139/149 (92%), Gaps = 1/149 (0%)
Frame = +2
Query: 547 ARLEAIRLLISFSVNHNIILHQMDVKSVFLNGYISEEVYVHQPPGFEDEKNPDHVFNLKK 606
++ EAIRLLISFSVNHNIILHQM+VKS FLNGYISEEVYVHQPPG EDEKN DH+F LKK
Sbjct: 86 SKTEAIRLLISFSVNHNIILHQMNVKSAFLNGYISEEVYVHQPPGXEDEKNSDHIFKLKK 265
Query: 607 SLYGLKQAPRAWYERL-RFLLENEFVRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANS 665
SLYGLKQAPRAWYERL FLLENE VRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSAN
Sbjct: 266 SLYGLKQAPRAWYERLSSFLLENEXVRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANP 445
Query: 666 SLCKEFSEMMQAEFEMSMMGELKYFLGIQ 694
SLCKEFSEMMQAEFEM MMGELKYFLGIQ
Sbjct: 446 SLCKEFSEMMQAEFEMRMMGELKYFLGIQ 532
Score = 35.4 bits (80), Expect(3) = 2e-77
Identities = 16/18 (88%), Positives = 16/18 (88%)
Frame = +3
Query: 531 YRQQEGIDYTETFAPVAR 548
Y QQ GIDYTETFAPVAR
Sbjct: 39 YSQQ*GIDYTETFAPVAR 92
Score = 23.1 bits (48), Expect(3) = 2e-77
Identities = 10/10 (100%), Positives = 10/10 (100%)
Frame = +1
Query: 521 RNKARLVAQG 530
RNKARLVAQG
Sbjct: 10 RNKARLVAQG 39
>BP084332
Length = 368
Score = 183 bits (465), Expect(2) = 1e-46
Identities = 94/112 (83%), Positives = 97/112 (85%)
Frame = +2
Query: 850 QSTIALSTAEAEYISIAICNT*MLWMKHQLEDYQILESNIPIYCDNTAAISLSKNPILHS 909
QSTIALSTAEAEYIS AIC+T MLWMKHQLEDYQILESNIPIYCDNTAAISLSKNPILHS
Sbjct: 32 QSTIALSTAEAEYISAAICSTQMLWMKHQLEDYQILESNIPIYCDNTAAISLSKNPILHS 211
Query: 910 RAKYIEVKYHFIRDYVQKGYFF*SSLILTINSADIFTKPLAEDRFKFILKNL 961
RAK+IEVKYHFIRDYVQKG + ADIFTKPLAEDRF FILKNL
Sbjct: 212 RAKHIEVKYHFIRDYVQKGVLLLKFVDTDHQWADIFTKPLAEDRFNFILKNL 367
Score = 21.6 bits (44), Expect(2) = 1e-46
Identities = 8/8 (100%), Positives = 8/8 (100%)
Frame = +1
Query: 835 CQFLGSNL 842
CQFLGSNL
Sbjct: 4 CQFLGSNL 27
>AV780009
Length = 529
Score = 117 bits (294), Expect = 7e-27
Identities = 61/163 (37%), Positives = 97/163 (59%), Gaps = 4/163 (2%)
Frame = -1
Query: 790 RILRYLKGTTNLGLLYKKTSEYKLSGYCDADYAGDRTERKSTSGNCQFLGSNLVSWASKR 849
R+LRY+KG GL + S KL Y D+D+AG R+S +G FLG++L+SW +K+
Sbjct: 517 RVLRYVKGAPAQGLFFSADSPLKLQAYSDSDWAGCPDTRRSVTGYSIFLGTSLISWRTKK 338
Query: 850 QSTIALSTAEAEYISIAICNT*MLWMKHQLEDYQILESN----IPIYCDNTAAISLSKNP 905
Q+T++ S++EAEY ++A + W+ + +Q L+ N +P++CDN +A+ ++ NP
Sbjct: 337 QTTVSRSSSEAEYRALAATVCEVQWLSYL---FQFLKLNVPLPVPLFCDNQSALHIAHNP 167
Query: 906 ILHSRAKYIEVKYHFIRDYVQKGYFF*SSLILTINSADIFTKP 948
H R K+IE+ H +R +Q G + ADIFTKP
Sbjct: 166 TFHERTKHIELDCHVVRAKLQAGLIHLLPISTHHQLADIFTKP 38
>BP052632
Length = 489
Score = 107 bits (267), Expect = 9e-24
Identities = 54/123 (43%), Positives = 78/123 (62%)
Frame = -3
Query: 293 KTPYELWKKVKPNISYFHPFGCVCYALNTKDRLHKFDSKSSKCLLLGYSKRSKGFRIYNT 352
KTPYELW+ +P +SYFHPFGC + LNT D + KFD+K K +LLGYS SK + ++N+
Sbjct: 487 KTPYELWRGRRPTVSYFHPFGCK*FILNTNDNIGKFDNKFDKRILLGYSSNSKAYIVFNS 308
Query: 353 DAKTIEEYIHVRFDDKLDSDQSKLVEKFVDMSINVSDKGKAPEEAEPEEDSPEEVGPSDP 412
+ +EE I+V+FDD +L + F+++ ++ D+ E +P E S EE PS
Sbjct: 307 RTQVVEESINVKFDD*SIEATYRLEKDFLNLDLSDQDEEVPTNEGQPSETSHEE--PS-T 137
Query: 413 QPQ 415
QPQ
Sbjct: 136 QPQ 128
>BP075943
Length = 547
Score = 65.1 bits (157), Expect = 5e-11
Identities = 41/159 (25%), Positives = 74/159 (45%), Gaps = 2/159 (1%)
Frame = -3
Query: 653 IYVDDIIFGSANSSLCKEFSEMMQAEFEMSMMGELKYFLGIQVDQTPEATYIHQSKYTKE 712
+YVDD++ + + + +F + +GE KYFLG+++ ++ ++Q KY +
Sbjct: 515 LYVDDVLLAGNDMHEIQLVKSSLHDQFRIKDLGEAKYFLGLEIARSTSGIVLNQRKYALQ 336
Query: 713 LLKKFNMTESIIAKTLMHPTCILEKEDASGKVCQKL--YRGMICSFLYLTASRPDILFSE 770
L+ + H T +G + YR ++ LYL +RPDI F+
Sbjct: 335 LISDSGHF-GFQPRFYSHGTNSQTLGTNTGTPLTDIGSYRRIVGRLLYLNTTRPDITFAV 159
Query: 771 HLCARFQSDPRETHLTVVKRILRYLKGTTNLGLLYKKTS 809
+ ++F S P + H + L+YL G+ GL Y +S
Sbjct: 158 NQLSQFLSAPTDIHEQQLTGFLKYL*GSPGSGLFYPASS 42
>AV766665
Length = 601
Score = 64.3 bits (155), Expect = 9e-11
Identities = 46/128 (35%), Positives = 65/128 (49%), Gaps = 4/128 (3%)
Frame = +3
Query: 738 EDASGKVCQKLYRGMICSFLYLTASRPDILF---SEHLCARFQSDPRETHL-TVVKRILR 793
E+A +C + +I ++YL IL SE L + R + L T RI
Sbjct: 207 EEAKIHLCDRGCVSLI-KYIYLKGDVSLILVRAQSEGLVQKTYGKNRWSSLKTFTTRIF* 383
Query: 794 YLKGTTNLGLLYKKTSEYKLSGYCDADYAGDRTERKSTSGNCQFLGSNLVSWASKRQSTI 853
YLK + G L++K + + G+ ADY G +R ST G FL NLV+W SK+Q+ I
Sbjct: 384 YLKANSRRGPLFQKEGKSSMDGFTYADYLGSIVDRLSTMGYYMFLSGNLVTWRSKQQNII 563
Query: 854 ALSTAEAE 861
A S+ EAE
Sbjct: 564 ARSSGEAE 587
>AV427921
Length = 284
Score = 64.3 bits (155), Expect = 9e-11
Identities = 34/80 (42%), Positives = 44/80 (54%)
Frame = +1
Query: 239 TPQQNGVVERKNRTLQEMSRTMLQEIDMAKHFWAEAVNTSCYIQNRISVRPILNKTPYEL 298
TPQQNGV ERKNRT+ M R++L + K F EAV S +I NR + N P E
Sbjct: 22 TPQQNGVSERKNRTILNMVRSLLTMSGLPKSFLPEAVMWSLHILNRSPTLVVQNMMPEEA 201
Query: 299 WKKVKPNISYFHPFGCVCYA 318
W +P + +F C+ YA
Sbjct: 202 WSGRQPAVDHFRISRCLAYA 261
>TC16929 weakly similar to UP|Q9LFY6 (Q9LFY6) T7N9.5, partial (4%)
Length = 553
Score = 62.8 bits (151), Expect = 3e-10
Identities = 30/91 (32%), Positives = 50/91 (53%), Gaps = 1/91 (1%)
Frame = +1
Query: 874 WMKHQLEDYQI-LESNIPIYCDNTAAISLSKNPILHSRAKYIEVKYHFIRDYVQKGYFF* 932
W+ + L+D ++ E +YCDN +A ++ NP+ H R K+IE+ H +R+ +QKG
Sbjct: 4 WLTYLLQDLKVPFEQPALVYCDNNSARHIAANPVFHERTKHIEIDCHIVRERIQKGLIHL 183
Query: 933 SSLILTINSADIFTKPLAEDRFKFILKNLNM 963
+ + ADI+TK L+ F I L +
Sbjct: 184 LPISSSEPLADIYTKALSPQNFHQICAKLGL 276
>BP063694
Length = 511
Score = 62.4 bits (150), Expect = 3e-10
Identities = 45/125 (36%), Positives = 60/125 (48%), Gaps = 3/125 (2%)
Frame = -2
Query: 78 EQWV*HRRLGHASMRKIS*LCK---LDLVRGLPTLKFSSDALCEACQKGKFTKVSFKAKN 134
E W H RLGH + I L K LD+ LP C +CQ K ++ F N
Sbjct: 363 ELW--HSRLGHVNFDIIKQLHKHGCLDVSSILP-----KPICCTSCQMAKSKRLVFHDNN 205
Query: 135 VVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYSRWTWVKFISRKDESHSVFSTFI 194
S L+L+H DL GP SI G Y ++ VDD+SR+TW + RK + V F
Sbjct: 204 K-RASAVLDLIHCDLRGPSPVASIDGFSYFVIFVDDFSRFTWFYPLKRKSDFSDVLLRFK 28
Query: 195 VQVQN 199
V ++N
Sbjct: 27 VFMEN 13
>TC19389 weakly similar to UP|POLX_TOBAC (P10978) Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
; Reverse transcriptase ; Endonuclease] , partial (6%)
Length = 498
Score = 42.0 bits (97), Expect(2) = 1e-09
Identities = 16/30 (53%), Positives = 23/30 (76%)
Frame = +2
Query: 891 IYCDNTAAISLSKNPILHSRAKYIEVKYHF 920
+YCDN AI L +N HSR+K+I+V+YH+
Sbjct: 173 LYCDNQGAIYLGQNSTFHSRSKHIDVRYHW 262
Score = 38.5 bits (88), Expect(2) = 1e-09
Identities = 18/39 (46%), Positives = 27/39 (69%)
Frame = +3
Query: 843 VSWASKRQSTIALSTAEAEYISIAICNT*MLWMKHQLED 881
V+W S+ Q +ALSTAEAE+I+ +LWMK+ L++
Sbjct: 27 VAWPSRLQKCVALSTAEAEFIAATEACHELLWMKNFLQN 143
>AU088980
Length = 360
Score = 60.1 bits (144), Expect = 2e-09
Identities = 28/115 (24%), Positives = 54/115 (46%)
Frame = +2
Query: 243 NGVVERKNRTLQEMSRTMLQEIDMAKHFWAEAVNTSCYIQNRISVRPILNKTPYELWKKV 302
N +VERK+R + ++R ++ + K+ W AV + Y+ N + + + PY+
Sbjct: 2 NSIVERKHRHILNVTRALMFHSYLPKNLWTFAVKHAAYLINXLPSPLLKGQCPYQFLNND 181
Query: 303 KPNISYFHPFGCVCYALNTKDRLHKFDSKSSKCLLLGYSKRSKGFRIYNTDAKTI 357
P + FG +CY+ K D ++ KC+ LG+ + G+ + + I
Sbjct: 182 SPTLLDLKVFGTLCYSSTLTHNXQKLDPRARKCVFLGFKTGTXGYIVMDLPTTXI 346
>BP057233
Length = 473
Score = 60.1 bits (144), Expect = 2e-09
Identities = 29/77 (37%), Positives = 46/77 (59%)
Frame = -2
Query: 891 IYCDNTAAISLSKNPILHSRAKYIEVKYHFIRDYVQKGYFF*SSLILTINSADIFTKPLA 950
++CDN +A +L+ NP+LH+R+K+IE+ H+IRD V + + + T AD TKPL+
Sbjct: 445 LWCDNLSAKALASNPVLHARSKHIEIDVHYIRDQVLQNEVVVAYVPTTDQIADCLTKPLS 266
Query: 951 EDRFKFILKNLNMDFCP 967
RF + L + P
Sbjct: 265 HTRFSQLRDKLGVIHSP 215
>BP043850
Length = 515
Score = 56.6 bits (135), Expect = 2e-08
Identities = 27/91 (29%), Positives = 49/91 (53%), Gaps = 1/91 (1%)
Frame = -1
Query: 872 MLWMKHQLEDYQILESN-IPIYCDNTAAISLSKNPILHSRAKYIEVKYHFIRDYVQKGYF 930
++W++ L + L+S P++ DNT+AI ++ NP+ H ++IEV H +R+ +
Sbjct: 509 IIWLRGLLSELGFLQSQPTPLHADNTSAIQIAANPVYHEWTRHIEVDCHSVREAYDRRVI 330
Query: 931 F*SSLILTINSADIFTKPLAEDRFKFILKNL 961
+ ++ ADI TK L R F++ L
Sbjct: 329 TLPHVSTSVQIADILTKSLTRQRHNFLVSKL 237
>TC10186 weakly similar to UP|Q9FH39 (Q9FH39) Copia-type polyprotein,
partial (4%)
Length = 528
Score = 38.5 bits (88), Expect(2) = 4e-07
Identities = 21/50 (42%), Positives = 27/50 (54%)
Frame = +3
Query: 914 IEVKYHFIRDYVQKGYFF*SSLILTINSADIFTKPLAEDRFKFILKNLNM 963
IE KYHF+RD V KG + ADI TK L +RF+ + LN+
Sbjct: 72 IETKYHFLRDQVTKGKISLKHCGTDLQVADIMTKGLKTERFRNMRAMLNV 221
Score = 33.1 bits (74), Expect(2) = 4e-07
Identities = 13/22 (59%), Positives = 18/22 (81%)
Frame = +2
Query: 894 DNTAAISLSKNPILHSRAKYIE 915
DN +AI L+KNP+ H R+K+IE
Sbjct: 5 DNKSAIDLAKNPVSHGRSKHIE 70
>TC15664 weakly similar to UP|O81617 (O81617) F8M12.17 protein, partial (4%)
Length = 670
Score = 51.6 bits (122), Expect = 6e-07
Identities = 25/91 (27%), Positives = 48/91 (52%), Gaps = 1/91 (1%)
Frame = +1
Query: 874 WMKHQLEDYQI-LESNIPIYCDNTAAISLSKNPILHSRAKYIEVKYHFIRDYVQKGYFF* 932
W+ + L+D ++ S +YCD+ +A ++ N + H R K++++ H +R+ +Q F
Sbjct: 13 WLTYILQDLRVPFISPSLLYCDSQSARHIATNAVFHERTKHLDIDCHVVREKLQAKLFHL 192
Query: 933 SSLILTINSADIFTKPLAEDRFKFILKNLNM 963
+ +ADI TKPL F ++ L +
Sbjct: 193 LPISSVDQTADILTKPLESGPFSHLVSKLGV 285
>TC12832 weakly similar to UP|POLX_TOBAC (P10978) Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
; Reverse transcriptase ; Endonuclease] , partial (9%)
Length = 747
Score = 50.4 bits (119), Expect = 1e-06
Identities = 27/101 (26%), Positives = 52/101 (50%), Gaps = 3/101 (2%)
Frame = +2
Query: 624 FLLENEFVRGKVDTTLFCKTYKD-DILIVQIYVDDIIFGSANSSLCKEFSEMMQAEFEMS 682
F++ + R D + K + D D +I+ +YVDD++ N +E + EF+M
Sbjct: 11 FIMSLGYNRLSSDHCTYHKRFDDNDFIILLLYVDDMLVVGPNKDRVQELKAQLAREFDMK 190
Query: 683 MMGELKYFLGIQV--DQTPEATYIHQSKYTKELLKKFNMTE 721
+G LG+Q+ D+ ++ Q Y +++L++FNM +
Sbjct: 191 DLGPANKILGMQIHRDRKDRRIWLSQKNYLQKVLRRFNMQD 313
>TC17418 similar to UP|Q94AX2 (Q94AX2) AT5g39790/MKM21_80, partial (20%)
Length = 739
Score = 47.4 bits (111), Expect = 1e-05
Identities = 33/99 (33%), Positives = 49/99 (49%), Gaps = 1/99 (1%)
Frame = -1
Query: 706 QSKYTKELLKKFNMTES-IIAKTLMHPTCILEKEDASGKVCQKLYRGMICSFLYLTASRP 764
Q Y ++LKKF MT S I+ T+ + + +V Y+ +I S YL R
Sbjct: 739 QKIYVDDILKKFKMTNSKYISTTIGGKEIEAGRRNGGKRVDSTYYKSLIGSVRYLNTVRS 560
Query: 765 DILFSEHLCARFQSDPRETHLTVVKRILRYLKGTTNLGL 803
DI+ L +RF +P + H +R LRY+KGT G+
Sbjct: 559 DIVCGVGLRSRFM-EP*DCH*QGAQRSLRYIKGTLKDGI 446
>AV424544
Length = 276
Score = 45.8 bits (107), Expect = 3e-05
Identities = 21/80 (26%), Positives = 42/80 (52%)
Frame = +3
Query: 630 FVRGKVDTTLFCKTYKDDILIVQIYVDDIIFGSANSSLCKEFSEMMQAEFEMSMMGELKY 689
+++ D +LF K ++ +YVDD+I + + + + +F + +G LKY
Sbjct: 36 YIQSAHDHSLFTKFRDASFTVILVYVDDLILAGNDLNEIQCVKNKLDIQFRIKDLGTLKY 215
Query: 690 FLGIQVDQTPEATYIHQSKY 709
FLG++V ++ ++ Q KY
Sbjct: 216 FLGLEVARSSCGLFLSQRKY 275
>TC8952 similar to UP|O82607 (O82607) T2L5.9 protein, partial (3%)
Length = 550
Score = 45.4 bits (106), Expect = 4e-05
Identities = 25/80 (31%), Positives = 42/80 (52%), Gaps = 1/80 (1%)
Frame = +3
Query: 873 LWMKHQLEDYQILESN-IPIYCDNTAAISLSKNPILHSRAKYIEVKYHFIRDYVQKGYFF 931
LW+ + L D +I + IYCDN +A+ L+ N + H R + IE+ H + V G
Sbjct: 27 LWLTYALADLRIASLLLVVIYCDNRSALHLAANSVFHKRTENIEIDCHIV*VKVLFGILH 206
Query: 932 *SSLILTINSADIFTKPLAE 951
+ + AD+FTK +++
Sbjct: 207 LLHVPSSDQVADVFTKTISQ 266
>BP033602
Length = 533
Score = 43.5 bits (101), Expect = 2e-04
Identities = 29/99 (29%), Positives = 51/99 (51%)
Frame = +3
Query: 74 SVNEEQWV*HRRLGHASMRKIS*LCKLDLVRGLPTLKFSSDALCEACQKGKFTKVSFKAK 133
SV ++ + H RLGH + + + L DL + + S CE+C K + + ++
Sbjct: 255 SVRDQIMLWHNRLGHPNFQYLRHLFP-DLFKNVNC----SSLECESCVLAKNQRAPYYSQ 419
Query: 134 NVVSTSRPLELLHIDLFGPVKTESIGGKRYGMVIVDDYS 172
SRP L+H D++GP K + GKR+ + +DD++
Sbjct: 420 PY-HASRPFYLIHSDVWGPSKITTQFGKRWFVTFIDDHT 533
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.322 0.137 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,200,038
Number of Sequences: 28460
Number of extensions: 223089
Number of successful extensions: 1024
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 1012
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 1019
length of query: 967
length of database: 4,897,600
effective HSP length: 99
effective length of query: 868
effective length of database: 2,080,060
effective search space: 1805492080
effective search space used: 1805492080
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0101.5


