

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0101.4
(376 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
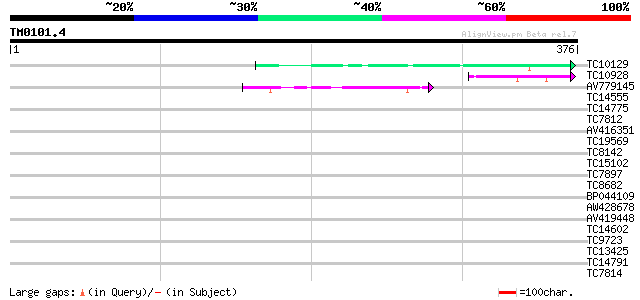
Score E
Sequences producing significant alignments: (bits) Value
TC10129 weakly similar to UP|Q7T9Y7 (Q7T9Y7) Odv-e66, partial (10%) 50 5e-07
TC10928 weakly similar to PIR|G96668|G96668 protein F1N19.7 [imp... 40 5e-04
AV779145 40 8e-04
TC14555 similar to UP|Q43558 (Q43558) Proline rich protein precu... 39 0.001
TC14775 PIR|S14181|S14181 DNA-directed RNA polymerase largest c... 38 0.002
TC7812 UP|Q9LKJ6 (Q9LKJ6) Water-selective transport intrinsic me... 38 0.003
AV416351 38 0.003
TC19569 similar to UP|AAR11301 (AAR11301) Lectin-like receptor k... 38 0.003
TC8142 UP|BAD11133 (BAD11133) Glutamate-rich protein, complete 37 0.004
TC15102 similar to UP|Q7X9B3 (Q7X9B3) 9/13 hydroperoxide lyase, ... 37 0.004
TC7897 weakly similar to UP|ENL2_ARATH (Q9T076) Early nodulin-li... 37 0.005
TC8682 similar to UP|HPPD_SOLSC (Q9ARF9) 4-hydroxyphenylpyruvate... 37 0.005
BP044109 36 0.009
AW428678 36 0.012
AV419448 36 0.012
TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinas... 35 0.016
TC9723 similar to AAQ65179 (AAQ65179) At3g10960, partial (25%) 35 0.016
TC13425 weakly similar to UP|Q7XJL5 (Q7XJL5) At2g17100 protein, ... 35 0.020
TC14791 similar to UP|Q9LJ04 (Q9LJ04) ESTs AU082210(C53655), par... 35 0.020
TC7814 similar to UP|Q9LKJ6 (Q9LKJ6) Water-selective transport i... 35 0.027
>TC10129 weakly similar to UP|Q7T9Y7 (Q7T9Y7) Odv-e66, partial (10%)
Length = 562
Score = 50.4 bits (119), Expect = 5e-07
Identities = 51/216 (23%), Positives = 77/216 (35%), Gaps = 4/216 (1%)
Frame = +1
Query: 164 PTPVDQPTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQTSEPIVSET 223
P+PV P S P+PP+ PS P P P+T P S
Sbjct: 1 PSPVYTPPSIPTPPKT--------------------PSPGNQPPHTPIPPKTPSPSYS-- 114
Query: 224 QAQEIPNPEKSNSEPQPSDHSNEEPSEREVLSPAPTVSFLTNVLFSSSFDKSEASRQFFQ 283
+P+P K+ P P++H P+ + SP ++ +
Sbjct: 115 -PPNVPSPPKA---PSPNNHPPYTPTPPKTPSPTSQPPYIP---LPPKTPSPISQPPHVP 273
Query: 284 LARERVSEILEHFLTVLSPNRYSGPRPEPLVDPDFLIQAVPLASMPHPPPPPRSPLPENN 343
S I + T +P + P +P P P++ P+ P PP++P P N
Sbjct: 274 TPPSTPSPISQPPYTP-TPPKTPSPTSQPPHTPTPPNTPSPISQPPYTPTPPKTPSPTNQ 450
Query: 344 ----ASVSNHSSDKSQNPETETPHQNPKTNTSAHQT 375
S N SS SQ P T +P + P + T
Sbjct: 451 PPHIPSPPNSSSPTSQPPNTPSPPKTPSPTSQPPNT 558
Score = 29.6 bits (65), Expect = 0.86
Identities = 21/82 (25%), Positives = 27/82 (32%)
Frame = +1
Query: 162 PEPTPVDQPTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQTSEPIVS 221
P P PTS P PS +P PS + P +P P +S P
Sbjct: 319 PTPPKTPSPTSQPPHTPTPPNTPSPISQPPYTPTPPKTPSPTNQPPHIPSPPNSSSPTSQ 498
Query: 222 ETQAQEIPNPEKSNSEPQPSDH 243
P S+P P+ H
Sbjct: 499 PPNTPSPPKTPSPTSQP-PNTH 561
>TC10928 weakly similar to PIR|G96668|G96668 protein F1N19.7 [imported] -
Arabidopsis thaliana {Arabidopsis thaliana;}, partial
(29%)
Length = 590
Score = 40.4 bits (93), Expect = 5e-04
Identities = 26/77 (33%), Positives = 35/77 (44%), Gaps = 6/77 (7%)
Frame = -3
Query: 305 YSGPRPEPLVDPDFLIQAVPLASMPHPPPPP----RSPLPENNASVSNHSSDKS--QNPE 358
+S P P P P+ P A+ P PPPPP SPL E + H+ P
Sbjct: 342 HSAP-PTPPTRPNTSPSQTPSAAAPPPPPPPPPAAPSPLSETRSGAPRHARPPPLYAPPA 166
Query: 359 TETPHQNPKTNTSAHQT 375
+P NP+T++S H T
Sbjct: 165 PPSPGCNPRTSSSLHAT 115
>AV779145
Length = 417
Score = 39.7 bits (91), Expect = 8e-04
Identities = 39/132 (29%), Positives = 59/132 (44%), Gaps = 5/132 (3%)
Frame = +2
Query: 155 AIYDETNPEPTPVDQPT--SHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYN 212
A + + P P+P QPT H +PP A+ P L A P+A++ P+ P +
Sbjct: 74 ARFSHSKPPPSPTTQPTPSPHGTPPHAT--------APGTASLAA--PAATSPPLTSPLS 223
Query: 213 PQTSEPIVSETQAQEIPNPEKSNSEPQPSDHSNEEP-SEREVLSPAPTVSF--LTNVLFS 269
P S + P+P +S P PS + P + LS ++ F L+N LF+
Sbjct: 224 P-------SPANSPPTPSPPSLSSPPSPSPTTRCAPATSHPRLSSLTSLRFLNLSNNLFN 382
Query: 270 SSFDKSEASRQF 281
+F S SR F
Sbjct: 383 GTF-PSHLSRLF 415
>TC14555 similar to UP|Q43558 (Q43558) Proline rich protein precursor,
partial (43%)
Length = 1038
Score = 39.3 bits (90), Expect = 0.001
Identities = 23/94 (24%), Positives = 40/94 (42%)
Frame = +2
Query: 165 TPVDQPTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQTSEPIVSETQ 224
+P + PT+ P+PP + QP + + P+ S+ P + P S P +++
Sbjct: 188 SPSNSPTTSPAPPTPTTPQPPAAQS-------SPPPAQSSPPPVQSSPPPASTPPPAQSS 346
Query: 225 AQEIPNPEKSNSEPQPSDHSNEEPSEREVLSPAP 258
+ +P S P P+ S P+ SP P
Sbjct: 347 PPPVSSPPPVQSTPPPAPASTPPPASPPPFSPPP 448
Score = 30.8 bits (68), Expect = 0.38
Identities = 28/123 (22%), Positives = 45/123 (35%)
Frame = +2
Query: 159 ETNPEPTPVDQPTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQTSEP 218
+++P P P PP + P P P A+T P L P TS
Sbjct: 338 QSSPPPVSSPPPVQSTPPPAPASTPPPASPPPFSPPPATPPPPAATPPPALTPVPATSPA 517
Query: 219 IVSETQAQEIPNPEKSNSEPQPSDHSNEEPSEREVLSPAPTVSFLTNVLFSSSFDKSEAS 278
+ P P + P + + PSE +P P++S L+ L ++ D S
Sbjct: 518 PAPAKVKSKSPAPAPA---PALAPVVSLSPSE----APGPSLSSLSPALSPAASDDSGVE 676
Query: 279 RQF 281
+ +
Sbjct: 677 KSW 685
Score = 28.1 bits (61), Expect = 2.5
Identities = 29/105 (27%), Positives = 41/105 (38%), Gaps = 5/105 (4%)
Frame = +2
Query: 159 ETNPEPTPVDQPTSHPSPPRASFFQPSIEEEPLCKM---LQAGKP--SASTSPIILPYNP 213
+++P P P SPP AS P+ P +Q+ P AST P P P
Sbjct: 257 QSSPPPAQSSPPPVQSSPPPASTPPPAQSSPPPVSSPPPVQSTPPPAPASTPPPASP--P 430
Query: 214 QTSEPIVSETQAQEIPNPEKSNSEPQPSDHSNEEPSEREVLSPAP 258
S P + P P + P P+ P++ + SPAP
Sbjct: 431 PFSPPPATPPPPAATPPPALT---PVPATSPAPAPAKVKSKSPAP 556
>TC14775 PIR|S14181|S14181 DNA-directed RNA polymerase largest chain
(isoform B1) - soybean (fragment) {Glycine
max;} , partial (22%)
Length = 848
Score = 38.1 bits (87), Expect = 0.002
Identities = 30/107 (28%), Positives = 44/107 (41%), Gaps = 13/107 (12%)
Frame = +1
Query: 157 YDETNPEPTPVD---QPTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNP 213
Y T+P +P PTS P+++ + PS+ P L P + TSP P +P
Sbjct: 49 YSPTSPSYSPTSPSYSPTSPSYNPQSAKYSPSLAYSPSSPRLSPSSPYSPTSPNYSPTSP 228
Query: 214 QTS--EPIVSETQAQEIP--------NPEKSNSEPQPSDHSNEEPSE 250
S P S + P +P+ S S PQ S + PS+
Sbjct: 229 SYSPTSPSYSPSSPTYSPSSPYNSGVSPDYSPSSPQFSPSTGYSPSQ 369
>TC7812 UP|Q9LKJ6 (Q9LKJ6) Water-selective transport intrinsic membrane
protein 1, complete
Length = 1149
Score = 37.7 bits (86), Expect = 0.003
Identities = 21/65 (32%), Positives = 31/65 (47%)
Frame = +1
Query: 195 LQAGKPSASTSPIILPYNPQTSEPIVSETQAQEIPNPEKSNSEPQPSDHSNEEPSEREVL 254
L + PS+S+SP +P +P T+ P + P P S+ P P+ + PS
Sbjct: 148 LSSSPPSSSSSPAPVPESPTTNSPTTAP------PPPPASSPPPSPTHSPSSSPSPSAPT 309
Query: 255 SPAPT 259
SPA T
Sbjct: 310 SPAAT 324
Score = 35.4 bits (80), Expect = 0.016
Identities = 28/93 (30%), Positives = 34/93 (36%)
Frame = +1
Query: 162 PEPTPVDQPTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQTSEPIVS 221
PE + PT+ P PP AS PS P + PSA TSP P S +
Sbjct: 193 PESPTTNSPTTAPPPPPASSPPPSPTHSP----SSSPSPSAPTSPAATSTPPSPSASSSA 360
Query: 222 ETQAQEIPNPEKSNSEPQPSDHSNEEPSEREVL 254
T + S S PS PS + L
Sbjct: 361 VTSPSSAVSSTSSLSFSDPSSPPCSSPSSPDSL 459
Score = 28.5 bits (62), Expect = 1.9
Identities = 20/78 (25%), Positives = 34/78 (42%)
Frame = +1
Query: 160 TNPEPTPVDQPTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQTSEPI 219
++P P+P P+S PSP + P+ P PSAS+S + P + +S
Sbjct: 247 SSPPPSPTHSPSSSPSPSAPT--SPAATSTP-------PSPSASSSAVTSPSSAVSSTSS 399
Query: 220 VSETQAQEIPNPEKSNSE 237
+S + P S+ +
Sbjct: 400 LSFSDPSSPPCSSPSSPD 453
Score = 28.1 bits (61), Expect = 2.5
Identities = 25/82 (30%), Positives = 35/82 (42%), Gaps = 5/82 (6%)
Frame = +1
Query: 296 FLTVLSPNRYSGPRPEPLVDPDFLIQAVPLASMPH--PPPPPRS---PLPENNASVSNHS 350
+L+ P+ S P P P P + P PPPPP S P P ++ S S
Sbjct: 145 WLSSSPPSSSSSPAPVP---------ESPTTNSPTTAPPPPPASSPPPSPTHSPSSSPSP 297
Query: 351 SDKSQNPETETPHQNPKTNTSA 372
S + T TP +P ++SA
Sbjct: 298 SAPTSPAATSTP-PSPSASSSA 360
>AV416351
Length = 428
Score = 37.7 bits (86), Expect = 0.003
Identities = 26/71 (36%), Positives = 33/71 (45%), Gaps = 1/71 (1%)
Frame = +3
Query: 299 VLSPNRYSGPRPEPLVDPDFLIQAVPLASMPHPPPPPRSPL-PENNASVSNHSSDKSQNP 357
++S + S P P+ P F + P P PPPPP S PE S NH
Sbjct: 21 IVSTSLPSSPLPKA---PPFSSSSPPRNPRPPPPPPPSSSSSPEKKNSPINHVLP----- 176
Query: 358 ETETPHQNPKT 368
+ TPHQ+PKT
Sbjct: 177 -SPTPHQSPKT 206
Score = 26.6 bits (57), Expect = 7.2
Identities = 27/104 (25%), Positives = 38/104 (35%), Gaps = 19/104 (18%)
Frame = +3
Query: 161 NPEPTPVDQPTSHPSP-------------PRASFFQPSIEEEPLCKM------LQAGKPS 201
NP P P P+S SP P +I + L M L A PS
Sbjct: 93 NPRPPPPPPPSSSSSPEKKNSPINHVLPSPTPHQSPKTIRQSDLSPMNSPKSKLSARPPS 272
Query: 202 ASTSPIILPYNPQTSEPIVSETQAQEIPNPEKSNSEPQPSDHSN 245
T P P +T EP V + + +K+ +P+ H+N
Sbjct: 273 PLTLP---PSQLKTEEPNVPDEAESKAVLVQKTIDKPKAWHHNN 395
>TC19569 similar to UP|AAR11301 (AAR11301) Lectin-like receptor kinase 1;1,
partial (16%)
Length = 486
Score = 37.7 bits (86), Expect = 0.003
Identities = 30/104 (28%), Positives = 38/104 (35%), Gaps = 6/104 (5%)
Frame = +1
Query: 161 NPEPTPVDQPTSHPSPPRASFFQPS------IEEEPLCKMLQAGKPSASTSPIILPYNPQ 214
+P P P QPT PS S P E P P++S S P NP
Sbjct: 70 SPPPPPSQQPTHSPSTSPTSTTTPPPSPTKVTENPPTAPSTSTKSPTSSASEEPSPPNPS 249
Query: 215 TSEPIVSETQAQEIPNPEKSNSEPQPSDHSNEEPSEREVLSPAP 258
TS T Q +P + P PS S + P+ S +P
Sbjct: 250 TS------TTQQPKHSPTSPPASPSPSPPSTKPPTAMASPSTSP 363
Score = 28.1 bits (61), Expect = 2.5
Identities = 22/72 (30%), Positives = 30/72 (41%), Gaps = 1/72 (1%)
Frame = +1
Query: 301 SPNRYSGPRPEPLVDPDFLIQAVPLASMPHPPPPPR-SPLPENNASVSNHSSDKSQNPET 359
S + YS P P P P P ++ PP P + + P S S S S + E
Sbjct: 55 SSSSYSPP-PPPSQQPTHSPSTSPTSTTTPPPSPTKVTENPPTAPSTSTKSPTSSASEEP 231
Query: 360 ETPHQNPKTNTS 371
P NP T+T+
Sbjct: 232 SPP--NPSTSTT 261
Score = 26.2 bits (56), Expect = 9.5
Identities = 28/127 (22%), Positives = 41/127 (32%), Gaps = 3/127 (2%)
Frame = +1
Query: 81 ASEEKDPSDGDDDDDDEPPPQKKGKKVRIATQVTHLQPTQELARTAPSTRITRSVVRASS 140
++ K P+ ++ P P TQ PT A +PS T+ +S
Sbjct: 187 STSTKSPTSSASEEPSPPNPSTS------TTQQPKHSPTSPPASPSPSPPSTKPPTAMAS 348
Query: 141 KFVVLLDDDLNLLDAIYDETNPEPTPVDQPTSHPSPPRASFFQPSIEEEP---LCKMLQA 197
P +P+ SHP+PP A PS P +
Sbjct: 349 ---------------------PSTSPLSATKSHPTPPAA----PSASSTPPPTTTSLKTM 453
Query: 198 GKPSAST 204
PS+ST
Sbjct: 454 SSPSSST 474
>TC8142 UP|BAD11133 (BAD11133) Glutamate-rich protein, complete
Length = 999
Score = 37.4 bits (85), Expect = 0.004
Identities = 30/107 (28%), Positives = 44/107 (41%), Gaps = 4/107 (3%)
Frame = +1
Query: 159 ETNPEPT-PVDQPTSHPSPPRASFFQPS--IEEEPLCKMLQAGKPSASTSPIILPYNPQT 215
ET PEP P + P P A P+ EEP K +P + + P P+T
Sbjct: 166 ETIPEPEQPAATEVAAPEQPAAEVPAPAEPATEEP--KEETTEEPKETETETEAPVAPET 339
Query: 216 SEPI-VSETQAQEIPNPEKSNSEPQPSDHSNEEPSEREVLSPAPTVS 261
+P+ V + E PE+ E + ++ EEP E + T S
Sbjct: 340 QDPLEVENKEVTEEAKPEEVKPETEKTEEKTEEPKTEEPAATTETES 480
>TC15102 similar to UP|Q7X9B3 (Q7X9B3) 9/13 hydroperoxide lyase, partial
(68%)
Length = 1097
Score = 37.4 bits (85), Expect = 0.004
Identities = 39/146 (26%), Positives = 58/146 (39%), Gaps = 2/146 (1%)
Frame = +1
Query: 118 PTQELARTAPSTRITRSVVRASSKFVVLLDDDLNLLDAIYDETNPEPTPVDQPTSHPSPP 177
P+ E TA +T T++ +S + + +P PTP +S P P
Sbjct: 157 PSSEPCTTATTTSTTKAATNSSPPESRSTTPPSSEPTCLLAPASP-PTPKSSHSSTPPPS 333
Query: 178 RASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQTSEPIVSETQAQEIPNPEKSNSE 237
R+S P K A +A + P P +P + P S+T PNP ++S+
Sbjct: 334 RSSSTTP--------KSKNATSLTAPSCP--PPASPAATAPAPSKTP----PNPPTNSSK 471
Query: 238 PQPSDHS--NEEPSEREVLSPAPTVS 261
P S N PS P+PT S
Sbjct: 472 PSSCKSSLPNTTPSSLSSAPPSPTTS 549
>TC7897 weakly similar to UP|ENL2_ARATH (Q9T076) Early nodulin-like protein
2 precursor (Phytocyanin-like protein), partial (31%)
Length = 1482
Score = 37.0 bits (84), Expect = 0.005
Identities = 28/104 (26%), Positives = 39/104 (36%), Gaps = 4/104 (3%)
Frame = +3
Query: 159 ETNPEPTPVDQPTSHPSP----PRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQ 214
+++P P+P P S P P + PS+ P + AG PSA T P
Sbjct: 549 KSSPSPSPKASPPSGSPPTAGEPSPAGTPPSVLPSPAGAPVPAGGPSAGTPP------SA 710
Query: 215 TSEPIVSETQAQEIPNPEKSNSEPQPSDHSNEEPSEREVLSPAP 258
++ P S A +P P S SN SP+P
Sbjct: 711 STSPSPSPVSAPPAASPAPGAESPSSSPTSNSPAGGPTATSPSP 842
>TC8682 similar to UP|HPPD_SOLSC (Q9ARF9) 4-hydroxyphenylpyruvate
dioxygenase (4HPPD) (HPD) (HPPDase) , partial (29%)
Length = 576
Score = 37.0 bits (84), Expect = 0.005
Identities = 26/73 (35%), Positives = 30/73 (40%), Gaps = 5/73 (6%)
Frame = +1
Query: 298 TVLSPNRYSGPRPEPLVDPDFLIQAVPLASMPHPP-----PPPRSPLPENNASVSNHSSD 352
T L+P S P P + P S P PP PPP SP P AS S S
Sbjct: 358 TTLTPPTSSAPATSASSSPPHTLPKSP--SPPPPPSPLSPPPPASPSPPPTASPSAPSPS 531
Query: 353 KSQNPETETPHQN 365
KS P +P Q+
Sbjct: 532 KSPTPSKPSPPQS 570
>BP044109
Length = 497
Score = 36.2 bits (82), Expect = 0.009
Identities = 18/46 (39%), Positives = 25/46 (54%), Gaps = 2/46 (4%)
Frame = +2
Query: 329 PHPPPPPRSPLPENNASVSNHSSD--KSQNPETETPHQNPKTNTSA 372
P PPPPP P P N + +H+S+ P T+TP P + TS+
Sbjct: 104 PSPPPPP--PPPNKNKTTPSHASNATSKSTPRTQTPTTPPPSPTSS 235
Score = 34.7 bits (78), Expect = 0.027
Identities = 36/125 (28%), Positives = 49/125 (38%), Gaps = 6/125 (4%)
Frame = +2
Query: 160 TNPEPTPVDQPTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQTSEPI 219
T TP PTS P PP ++F A KP STSP P P +S
Sbjct: 194 TQTPTTPPPSPTSSPKPPPSTF---------------ATKP--STSP---PETPSSS--- 304
Query: 220 VSETQAQEIPNPEKSN------SEPQPSDHSNEEPSEREVLSPAPTVSFLTNVLFSSSFD 273
S A +P+P S+ S P P + S R +PAP++ + +S
Sbjct: 305 -SHGPAPTLPSPPSSSTPTSTPSPPSPPNGSTLPSPPRATPTPAPSLPAARRMTSASLCS 481
Query: 274 KSEAS 278
S+ S
Sbjct: 482 TSKPS 496
>AW428678
Length = 394
Score = 35.8 bits (81), Expect = 0.012
Identities = 25/76 (32%), Positives = 32/76 (41%), Gaps = 8/76 (10%)
Frame = +2
Query: 306 SGPRPEPLVDPDFLIQAVPLASMPHPP------PPPRS--PLPENNASVSNHSSDKSQNP 357
+ P P P P + P + +PHPP PPP S P P + S S S P
Sbjct: 14 ASPSPPP---PYYYKSPPPPSPIPHPPYYYKSPPPPTSSPPPPYHYVSPPPPSPSSSTTP 184
Query: 358 ETETPHQNPKTNTSAH 373
+T T NP T +H
Sbjct: 185 KTTTNKSNPNTARLSH 232
>AV419448
Length = 382
Score = 35.8 bits (81), Expect = 0.012
Identities = 31/115 (26%), Positives = 43/115 (36%)
Frame = +2
Query: 231 PEKSNSEPQPSDHSNEEPSEREVLSPAPTVSFLTNVLFSSSFDKSEASRQFFQLARERVS 290
P S+ P PS S P P+PT S T S+S + +S
Sbjct: 32 PSSSHPPPSPSQPSLPSPPLHPPTPPSPTASSWT----SASAPPPSSPTATPSAPTPPLS 199
Query: 291 EILEHFLTVLSPNRYSGPRPEPLVDPDFLIQAVPLASMPHPPPPPRSPLPENNAS 345
L L+ S P P P+ +P AS+P PPPP + P + +S
Sbjct: 200 AALS-----LASTATSSPSPSPISNPS--------ASLPPTPPPPPTKTPSSTSS 325
Score = 30.0 bits (66), Expect = 0.66
Identities = 26/97 (26%), Positives = 37/97 (37%), Gaps = 1/97 (1%)
Frame = +2
Query: 156 IYDETNPEPTPVDQPTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPY-NPQ 214
++ T P PT ++ PP + PS PL L +A++SP P NP
Sbjct: 89 LHPPTPPSPTASSWTSASAPPPSSPTATPSAPTPPLSAALSLAS-TATSSPSPSPISNPS 265
Query: 215 TSEPIVSETQAQEIPNPEKSNSEPQPSDHSNEEPSER 251
S P P P K+ S S + P+ R
Sbjct: 266 ASLPPTPP------PPPTKTPSSTSSSPAATSSPAAR 358
>TC14602 UP|Q8GS56 (Q8GS56) Phosphoenolpyruvate carboxylase kinase, complete
Length = 1256
Score = 35.4 bits (80), Expect = 0.016
Identities = 18/45 (40%), Positives = 22/45 (48%)
Frame = -1
Query: 329 PHPPPPPRSPLPENNASVSNHSSDKSQNPETETPHQNPKTNTSAH 373
PH P P + P N AS HSS S +T P+QNP + H
Sbjct: 605 PHTPSPSKPPAQHNTASQPPHSS--SSRHQTTPPNQNPPVSDLHH 477
>TC9723 similar to AAQ65179 (AAQ65179) At3g10960, partial (25%)
Length = 689
Score = 35.4 bits (80), Expect = 0.016
Identities = 25/80 (31%), Positives = 36/80 (44%), Gaps = 4/80 (5%)
Frame = +3
Query: 165 TPVDQPTSHPSPPRASFF---QPSIEEEPLCKMLQAGKPSASTS-PIILPYNPQTSEPIV 220
TP P+S P+PPR+S + PS L + A P+AS S P +LP +P P
Sbjct: 333 TPPSPPSSAPAPPRSSLWPTSSPSTPPSYLTPVEPALFPTASPSAPTLLPLSPPAPAPPS 512
Query: 221 SETQAQEIPNPEKSNSEPQP 240
+ N +S + P
Sbjct: 513 ASFSLTSPANSTQSTRDTNP 572
>TC13425 weakly similar to UP|Q7XJL5 (Q7XJL5) At2g17100 protein, partial
(6%)
Length = 538
Score = 35.0 bits (79), Expect = 0.020
Identities = 25/90 (27%), Positives = 37/90 (40%), Gaps = 13/90 (14%)
Frame = +1
Query: 276 EASRQFFQLARERVSEI-------------LEHFLTVLSPNRYSGPRPEPLVDPDFLIQA 322
EA RQ + LA V+ + +HF + + S P P PL
Sbjct: 139 EALRQSYALADAHVAHMDSLRTLGPALVCFFDHFQETKTVSILSSPPPPPL--------- 291
Query: 323 VPLASMPHPPPPPRSPLPENNASVSNHSSD 352
P+A+ PH PPP P ++ S +H +D
Sbjct: 292 -PVAAPPHSPPPCSDEFPLHSESDDDHDAD 378
>TC14791 similar to UP|Q9LJ04 (Q9LJ04) ESTs AU082210(C53655), partial (37%)
Length = 1779
Score = 35.0 bits (79), Expect = 0.020
Identities = 30/117 (25%), Positives = 40/117 (33%)
Frame = -2
Query: 164 PTPVDQPTSHPSPPRASFFQPSIEEEPLCKMLQAGKPSASTSPIILPYNPQTSEPIVSET 223
P P D P + +PS P CK +A PSAS P + T
Sbjct: 1127 PQPSDNTECALHIPCLALLKPSWHSPPTCKQ-KAQTPSASRCSYQAPRDHLQPPQTA*RT 951
Query: 224 QAQEIPNPEKSNSEPQPSDHSNEEPSEREVLSPAPTVSFLTNVLFSSSFDKSEASRQ 280
QA + S+ + S ++P E PAP T S SE S +
Sbjct: 950 QAASMVQYPSSHCSQRSSSGCTQKPEEHSAALPAPVQRMPTETRSFGSQSSSEDSSE 780
>TC7814 similar to UP|Q9LKJ6 (Q9LKJ6) Water-selective transport intrinsic
membrane protein 1, partial (37%)
Length = 660
Score = 34.7 bits (78), Expect = 0.027
Identities = 20/65 (30%), Positives = 29/65 (43%)
Frame = +2
Query: 195 LQAGKPSASTSPIILPYNPQTSEPIVSETQAQEIPNPEKSNSEPQPSDHSNEEPSEREVL 254
L + PS+S+SP +P +P + P+ P P S+ P P + PS
Sbjct: 452 LSSSPPSSSSSPAPVPESPTINSPMTVP------PPPPXSSPPPSPMHSPSSSPSXSSPT 613
Query: 255 SPAPT 259
SPA T
Sbjct: 614 SPAAT 628
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.309 0.129 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,095,241
Number of Sequences: 28460
Number of extensions: 141755
Number of successful extensions: 3547
Number of sequences better than 10.0: 440
Number of HSP's better than 10.0 without gapping: 2378
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3156
length of query: 376
length of database: 4,897,600
effective HSP length: 92
effective length of query: 284
effective length of database: 2,279,280
effective search space: 647315520
effective search space used: 647315520
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0101.4


