

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0100b.1
(354 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
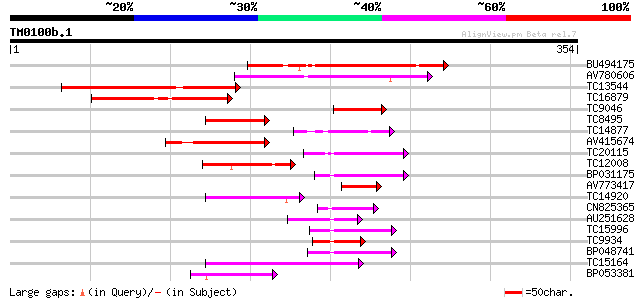
Score E
Sequences producing significant alignments: (bits) Value
BU494175 133 5e-32
AV780606 112 9e-26
TC13544 similar to UP|Q9LU42 (Q9LU42) Emb|CAB45082.1, partial (11%) 112 1e-25
TC16879 similar to UP|Q9LRH6 (Q9LRH6) ZIM, partial (17%) 73 6e-14
TC9046 homologue to UP|Q9LRH6 (Q9LRH6) ZIM, partial (10%) 67 3e-12
TC8495 similar to UP|Q9LVG4 (Q9LVG4) Emb|CAB86035.1 (Pseudo-resp... 52 2e-07
TC14877 similar to GB|AAP37701.1|30725358|BT008342 At4g32890 {Ar... 52 2e-07
AV415674 50 4e-07
TC20115 similar to PIR|T52104|T52104 GATA-binding transcription ... 49 1e-06
TC12008 similar to GB|BAB13743.1|10281006|AB046955 pseudo-respon... 49 1e-06
BP031175 48 3e-06
AV773417 47 4e-06
TC14920 similar to UP|BAC92733 (BAC92733) Hd1-like protein, part... 47 6e-06
CN825365 46 8e-06
AU251628 46 1e-05
TC15996 similar to UP|BAC98491 (BAC98491) AG-motif binding prote... 46 1e-05
TC9934 similar to UP|BAC98494 (BAC98494) AG-motif binding protei... 45 2e-05
BP048741 45 2e-05
TC15164 similar to UP|Q9ZTK7 (Q9ZTK7) CONSTANS-like protein 2, p... 44 4e-05
BP053381 43 9e-05
>BU494175
Length = 475
Score = 133 bits (334), Expect = 5e-32
Identities = 80/132 (60%), Positives = 90/132 (67%), Gaps = 6/132 (4%)
Frame = +2
Query: 149 KEVAQRMHRKNGQFASLKEDYKSPAGNLDSS-----NGTPPCPESIERRCQHCGTSEKST 203
KEVAQRMHRKNGQFASLKE S N DS+ NGTP ES+ RRCQHCG SE +T
Sbjct: 17 KEVAQRMHRKNGQFASLKEGPGS--SNWDSAQSSGQNGTPNS-ESL-RRCQHCGVSENNT 184
Query: 204 PAMRRGPAGPRSLCNACGLMWANKGTLRDLSKAAKT-PYEQNERDTSADMKPSTTEPENS 262
PAMRRGP GPR+LCNACGLMWANKGTLR L K ++ Q+E D S D+KP T P
Sbjct: 185 PAMRRGPDGPRTLCNACGLMWANKGTLRVLRKGGRSISVLQSEPDLSIDVKP-TVLPGEL 361
Query: 263 CADQAEEGTPEE 274
Q E+G E+
Sbjct: 362 PVIQDEQGISED 397
>AV780606
Length = 585
Score = 112 bits (280), Expect = 9e-26
Identities = 61/128 (47%), Positives = 73/128 (56%), Gaps = 4/128 (3%)
Frame = -1
Query: 141 KKIRYTCRKEVAQRMHRKNGQFASLKEDYKSPAGNLDSSNGTPPCPESIERRCQHCGTSE 200
KK+RY R+E+A RMHR GQ S K+ + D +G E C HCGTS
Sbjct: 585 KKVRYGVRQELALRMHRHKGQSTSSKKQDGANGSGTDQESGQDD--SQAETSCTHCGTSS 412
Query: 201 KSTPAMRRGPAGPRSLCNACGLMWANKGTLRDLSKA----AKTPYEQNERDTSADMKPST 256
KSTP MRRGP+GPRSLCNACGL WAN+G LRDLSK + EQ + SAD + +
Sbjct: 411 KSTPMMRRGPSGPRSLCNACGLFWANRGALRDLSKRNQEHSLVQVEQVDGGNSADCRTAM 232
Query: 257 TEPENSCA 264
N A
Sbjct: 231 PALNNVVA 208
>TC13544 similar to UP|Q9LU42 (Q9LU42) Emb|CAB45082.1, partial (11%)
Length = 579
Score = 112 bits (279), Expect = 1e-25
Identities = 66/114 (57%), Positives = 79/114 (68%), Gaps = 2/114 (1%)
Frame = +1
Query: 33 DEGQHAPATVAASASNASAPHP-RAGELTISFEGEVYVFPAVTPEKVQAVLLLLGQKDIP 91
+E P +VA SA++ + P R ELT+SFEGEVYVFPAVTP KVQAVLLLLG +D+
Sbjct: 250 EEADVNPVSVAVSANHEAVVLPSRTSELTLSFEGEVYVFPAVTPXKVQAVLLLLGGRDVQ 429
Query: 92 NSTPNSGL-LQQKNFPQIKGINDPSQSSNLSRRFASLVRFREKRKERCFEKKIR 144
S + L Q N +G + + SNLSRR ASLVRFREKRKERCF+KKIR
Sbjct: 430 ASMSTVEVPLDQSN----RGTDGNPKRSNLSRRIASLVRFREKRKERCFDKKIR 579
>TC16879 similar to UP|Q9LRH6 (Q9LRH6) ZIM, partial (17%)
Length = 622
Score = 73.2 bits (178), Expect = 6e-14
Identities = 43/88 (48%), Positives = 60/88 (67%)
Frame = +2
Query: 52 PHPRAGELTISFEGEVYVFPAVTPEKVQAVLLLLGQKDIPNSTPNSGLLQQKNFPQIKGI 111
P + +LT+SF G+VYVF +VTPEKVQAVLLLLG ++ S+ + GL + + +G
Sbjct: 371 PCDDSSQLTLSFRGQVYVFDSVTPEKVQAVLLLLGGCEL--SSGSQGL--EISPTNQRGS 538
Query: 112 NDPSQSSNLSRRFASLVRFREKRKERCF 139
+ +L +R ASL+RFR+KRKERCF
Sbjct: 539 AEFPAKCSLPQRAASLIRFRQKRKERCF 622
>TC9046 homologue to UP|Q9LRH6 (Q9LRH6) ZIM, partial (10%)
Length = 629
Score = 67.4 bits (163), Expect = 3e-12
Identities = 28/33 (84%), Positives = 31/33 (93%)
Frame = +3
Query: 203 TPAMRRGPAGPRSLCNACGLMWANKGTLRDLSK 235
TP MRRGP+GPRSLCNACGL WAN+GTLRDLS+
Sbjct: 3 TPMMRRGPSGPRSLCNACGLFWANRGTLRDLSR 101
>TC8495 similar to UP|Q9LVG4 (Q9LVG4) Emb|CAB86035.1 (Pseudo-response
regulator 3), partial (9%)
Length = 783
Score = 52.0 bits (123), Expect = 2e-07
Identities = 24/40 (60%), Positives = 31/40 (77%)
Frame = +1
Query: 123 RFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQF 162
R A+L +FR KRKERCFEKK+RY RK++A++ R GQF
Sbjct: 76 REAALAKFRLKRKERCFEKKVRYHSRKKLAEQRPRIRGQF 195
>TC14877 similar to GB|AAP37701.1|30725358|BT008342 At4g32890 {Arabidopsis
thaliana;}, partial (25%)
Length = 1039
Score = 52.0 bits (123), Expect = 2e-07
Identities = 28/63 (44%), Positives = 36/63 (56%)
Frame = +3
Query: 178 SSNGTPPCPESIERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTLRDLSKAA 237
S+NG P P RRC HC + TP R GP GP++LCNACG+ + G L + A
Sbjct: 441 SNNGQNPMP----RRCTHC--LSQRTPQWRAGPLGPKTLCNACGVRY-KSGRLHPEYRPA 599
Query: 238 KTP 240
K+P
Sbjct: 600 KSP 608
>AV415674
Length = 319
Score = 50.4 bits (119), Expect = 4e-07
Identities = 26/65 (40%), Positives = 39/65 (60%)
Frame = +2
Query: 98 GLLQQKNFPQIKGINDPSQSSNLSRRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHR 157
G++ +KN + + + R A+L +FR KRKERCFE K+RY RK++A++ R
Sbjct: 74 GVVDRKNTMNV------ADEERTALREAALAKFRLKRKERCFENKVRYHSRKKLAEQRPR 235
Query: 158 KNGQF 162
GQF
Sbjct: 236 IRGQF 250
>TC20115 similar to PIR|T52104|T52104 GATA-binding transcription factor
homolog 2 [imported] - Arabidopsis
thaliana {Arabidopsis thaliana;}, partial (31%)
Length = 635
Score = 49.3 bits (116), Expect = 1e-06
Identities = 26/66 (39%), Positives = 36/66 (54%)
Frame = +3
Query: 184 PCPESIERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTLRDLSKAAKTPYEQ 243
P E RRC HC SEK TP R GP GP++LCNACG+ + + + + AA +
Sbjct: 78 PAGEGGVRRCSHCA-SEK-TPQWRTGPLGPKTLCNACGVRFKSGRLVPEYRPAASPTFVL 251
Query: 244 NERDTS 249
++ S
Sbjct: 252 SQHSNS 269
>TC12008 similar to GB|BAB13743.1|10281006|AB046955 pseudo-response
regulator 5 {Arabidopsis thaliana;} , partial (7%)
Length = 496
Score = 48.9 bits (115), Expect = 1e-06
Identities = 27/61 (44%), Positives = 39/61 (63%), Gaps = 3/61 (4%)
Frame = +3
Query: 121 SRRFASLVRFREKRKER---CFEKKIRYTCRKEVAQRMHRKNGQFASLKEDYKSPAGNLD 177
S+R A+L +FR+KRKER CF KK+RY RK +A++ R GQF + ++ +G D
Sbjct: 96 SQRQAALTKFRQKRKEREERCFHKKVRYQSRKRLAEQRPRFRGQFVR-QSSNENASGATD 272
Query: 178 S 178
S
Sbjct: 273 S 275
>BP031175
Length = 537
Score = 47.8 bits (112), Expect = 3e-06
Identities = 22/59 (37%), Positives = 32/59 (53%)
Frame = -2
Query: 191 RRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTLRDLSKAAKTPYEQNERDTS 249
RRC HC T + TP R GP GP++LCNACG+ + + + + AA + + S
Sbjct: 524 RRCLHCATDK--TPQWRTGPMGPKTLCNACGVRYKSGRLVPEYRPAASPTFMLTKHSNS 354
>AV773417
Length = 411
Score = 47.4 bits (111), Expect = 4e-06
Identities = 19/25 (76%), Positives = 21/25 (84%)
Frame = -3
Query: 208 RGPAGPRSLCNACGLMWANKGTLRD 232
RGP G RSLC+ACGL WAN+G LRD
Sbjct: 409 RGPFGSRSLCHACGLFWANRGALRD 335
>TC14920 similar to UP|BAC92733 (BAC92733) Hd1-like protein, partial (15%)
Length = 645
Score = 46.6 bits (109), Expect = 6e-06
Identities = 27/65 (41%), Positives = 34/65 (51%), Gaps = 3/65 (4%)
Frame = +3
Query: 123 RFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQFASLKEDYKS---PAGNLDSS 179
R A ++R+REKRK R FEK IRY RK A+ R G+FA +D PA
Sbjct: 132 REAKVMRYREKRKNRRFEKTIRYASRKAYAESRPRIKGRFAKRSDDNNHADLPAAGATGY 311
Query: 180 NGTPP 184
+G P
Sbjct: 312 DGLVP 326
>CN825365
Length = 511
Score = 46.2 bits (108), Expect = 8e-06
Identities = 21/38 (55%), Positives = 22/38 (57%)
Frame = +3
Query: 193 CQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTL 230
C HCG + STP R GP LCNACG W KGTL
Sbjct: 165 CYHCGVT--STPLWRNGPPEKPVLCNACGSRWRTKGTL 272
>AU251628
Length = 350
Score = 45.8 bits (107), Expect = 1e-05
Identities = 22/47 (46%), Positives = 28/47 (58%)
Frame = +3
Query: 174 GNLDSSNGTPPCPESIERRCQHCGTSEKSTPAMRRGPAGPRSLCNAC 220
GN +S+ ++ C CGTS+ TP R GPAGP+SLCNAC
Sbjct: 216 GNPNSAAAAASSGSEQKKTCADCGTSK--TPLWRGGPAGPKSLCNAC 350
>TC15996 similar to UP|BAC98491 (BAC98491) AG-motif binding protein-1,
partial (19%)
Length = 555
Score = 45.8 bits (107), Expect = 1e-05
Identities = 19/54 (35%), Positives = 31/54 (57%)
Frame = +2
Query: 188 SIERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTLRDLSKAAKTPY 241
++ R+C HC ++ TP R GP GP++LCNACG+ + + + AA +
Sbjct: 152 AVPRKCMHCEVTK--TPQWREGPMGPKTLCNACGVRYRSGRLFPEYRPAASPTF 307
>TC9934 similar to UP|BAC98494 (BAC98494) AG-motif binding protein-4,
partial (37%)
Length = 1050
Score = 45.1 bits (105), Expect = 2e-05
Identities = 18/33 (54%), Positives = 23/33 (69%)
Frame = +2
Query: 190 ERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGL 222
+RRC HC + TP R GP GP++LCNACG+
Sbjct: 566 QRRCSHCQVQK--TPQWRAGPLGPKTLCNACGV 658
>BP048741
Length = 558
Score = 44.7 bits (104), Expect = 2e-05
Identities = 20/55 (36%), Positives = 30/55 (54%)
Frame = -1
Query: 187 ESIERRCQHCGTSEKSTPAMRRGPAGPRSLCNACGLMWANKGTLRDLSKAAKTPY 241
++ RRC HCG + TP R GP G ++LCNACG+ + + L + A +
Sbjct: 474 QAAPRRCSHCGVQK--TPQWRTGPHGAKTLCNACGVRFKSGRLLPEYRPACSPTF 316
>TC15164 similar to UP|Q9ZTK7 (Q9ZTK7) CONSTANS-like protein 2, partial
(57%)
Length = 1185
Score = 43.9 bits (102), Expect = 4e-05
Identities = 32/99 (32%), Positives = 44/99 (44%)
Frame = +1
Query: 123 RFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQFASLKEDYKSPAGNLDSSNGT 182
R A ++R+REKRK R FEK IRY RK A+ R G+FA + + D +G
Sbjct: 847 REARVLRYREKRKNRRFEKTIRYASRKAYAETRPRIKGRFAKRSDVNVNLLTEEDPFSGY 1026
Query: 183 PPCPESIERRCQHCGTSEKSTPAMRRGPAGPRSLCNACG 221
P + + S + MR G +ACG
Sbjct: 1027GVVPTC*MKAVVYVYMSGVRSSPMRMKVVGHLL*ASACG 1143
>BP053381
Length = 524
Score = 42.7 bits (99), Expect = 9e-05
Identities = 23/56 (41%), Positives = 32/56 (57%), Gaps = 2/56 (3%)
Frame = -2
Query: 114 PSQSSNLS--RRFASLVRFREKRKERCFEKKIRYTCRKEVAQRMHRKNGQFASLKE 167
P SSN + R L R+R K+ +R F +KI+Y CRK +A R G+FA +E
Sbjct: 445 PFFSSNCTSEERLQKLSRYRNKKTKRNFGRKIKYACRKALADSQPRIRGRFAKTEE 278
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.311 0.128 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,865,143
Number of Sequences: 28460
Number of extensions: 78132
Number of successful extensions: 400
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 389
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 393
length of query: 354
length of database: 4,897,600
effective HSP length: 91
effective length of query: 263
effective length of database: 2,307,740
effective search space: 606935620
effective search space used: 606935620
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 56 (26.2 bits)
Lotus: description of TM0100b.1


