

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0100a.4
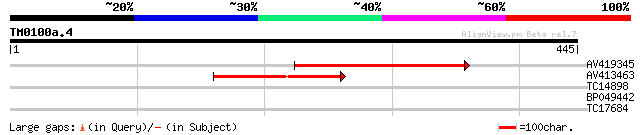
(445 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AV419345 264 2e-71
AV413463 118 2e-27
TC14898 32 0.16
BP049442 30 0.80
TC17684 27 6.8
>AV419345
Length = 416
Score = 264 bits (675), Expect = 2e-71
Identities = 125/138 (90%), Positives = 130/138 (93%)
Frame = +3
Query: 224 YIYPLYGLGELPQAFARLSAVYGGTYMLNKPECKVEFDSAGKAIGVTSEGETAKCKKVVC 283
YIYPLYGLGELPQ FARLSAVYGGTYMLNKPECKVEFD GK +GVTSEGETAKCKKVVC
Sbjct: 3 YIYPLYGLGELPQGFARLSAVYGGTYMLNKPECKVEFDEEGKVVGVTSEGETAKCKKVVC 182
Query: 284 DPSYLPDKVRNVGKVARAICIMSHPIPDTNDSHSAQVILPQKQLGRKSDMYLFCCSYAHN 343
DPSYLP KVR VGKVARAICIMSHPIP+TNDS S QVILPQKQLGRKSDMYLFCCSY+HN
Sbjct: 183 DPSYLPSKVRKVGKVARAICIMSHPIPNTNDSPSVQVILPQKQLGRKSDMYLFCCSYSHN 362
Query: 344 VAPKGKYIAFVTTEAETD 361
VAPKGK+IAFV+TEAETD
Sbjct: 363 VAPKGKFIAFVSTEAETD 416
>AV413463
Length = 309
Score = 118 bits (296), Expect = 2e-27
Identities = 62/103 (60%), Positives = 76/103 (73%)
Frame = +2
Query: 161 DLNQVTARQLISKYGLEDDTIDFIGHALALHLDDSYLDTPAKDFVERIKVYAESLARFQG 220
DL + L S +GL+ DTI F+GHALAL+ D+SYL PA + V ++++YA SLAR G
Sbjct: 2 DLTTLPMSGLYSHFGLQPDTIAFVGHALALYRDESYLSRPAVETVVKVQLYATSLAR-HG 178
Query: 221 GSPYIYPLYGLGELPQAFARLSAVYGGTYMLNKPECKVEFDSA 263
SPY+YPLYGLGELPQAFARLSAV+GGTYML P +V D A
Sbjct: 179 RSPYLYPLYGLGELPQAFARLSAVHGGTYMLATPVDEVLTDPA 307
>TC14898
Length = 582
Score = 32.3 bits (72), Expect = 0.16
Identities = 16/18 (88%), Positives = 17/18 (93%)
Frame = -1
Query: 428 KVLDLSVDLSAASATAEE 445
+VLDLSVDLSAASAT EE
Sbjct: 345 QVLDLSVDLSAASATTEE 292
>BP049442
Length = 494
Score = 30.0 bits (66), Expect = 0.80
Identities = 18/45 (40%), Positives = 25/45 (55%), Gaps = 1/45 (2%)
Frame = -2
Query: 5 YDVIVLGTGLKECILSGLLSVDGLKVLHM-DRNDYYGGASTSLNL 48
+DV V+GTGL + GLL D V+H+ +R +GG L L
Sbjct: 334 FDVSVVGTGLAVIL*LGLLGEDQRLVVHLHERGGSFGGDGEVLEL 200
>TC17684
Length = 721
Score = 26.9 bits (58), Expect = 6.8
Identities = 13/41 (31%), Positives = 24/41 (57%)
Frame = +1
Query: 6 DVIVLGTGLKECILSGLLSVDGLKVLHMDRNDYYGGASTSL 46
D +V+G+G +++G+L+ G KVL ++ Y + SL
Sbjct: 475 DAVVVGSGSGGGLIAGILAKAGYKVLVKEKGGYTARNNLSL 597
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.137 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,672,770
Number of Sequences: 28460
Number of extensions: 84570
Number of successful extensions: 303
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 302
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 302
length of query: 445
length of database: 4,897,600
effective HSP length: 93
effective length of query: 352
effective length of database: 2,250,820
effective search space: 792288640
effective search space used: 792288640
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0100a.4


