

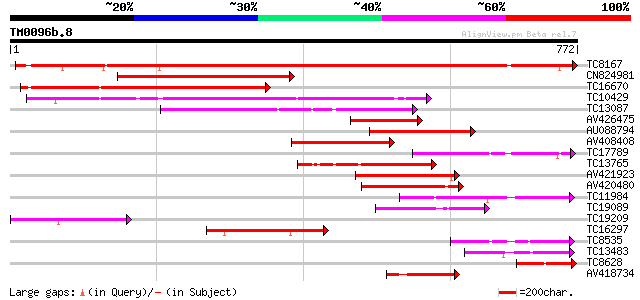
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0096b.8
(772 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC8167 similar to PIR|JC7519|JC7519 subtilisin-like serine prote... 734 0.0
CN824981 487 e-138
TC16670 similar to PIR|JC7519|JC7519 subtilisin-like serine prot... 451 e-127
TC10429 similar to UP|Q9ZR46 (Q9ZR46) P69C protein, partial (52%) 411 e-115
TC13087 similar to UP|Q9FIF8 (Q9FIF8) Serine protease-like prote... 241 2e-64
AV426475 201 3e-52
AU088794 187 7e-48
AV408408 185 3e-47
TC17789 similar to PIR|T05768|T05768 subtilisin-like proteinase ... 182 2e-46
TC13765 similar to UP|Q9LLL8 (Q9LLL8) Subtilisin-type serine end... 152 2e-37
AV421923 148 3e-36
AV420480 145 2e-35
TC11984 similar to UP|Q93WQ0 (Q93WQ0) Subtilisin-type protease, ... 144 7e-35
TC19089 similar to UP|Q9ZTT3 (Q9ZTT3) Subtilisin-like protease C... 131 5e-31
TC19209 similar to GB|BAB11244.1|10177874|AB010074 serine protea... 129 1e-30
TC16297 weakly similar to UP|Q93WQ0 (Q93WQ0) Subtilisin-type pro... 123 1e-28
TC8535 weakly similar to UP|Q9FJF3 (Q9FJF3) Serine protease-like... 108 4e-24
TC13483 weakly similar to UP|Q9FIF8 (Q9FIF8) Serine protease-lik... 94 6e-20
TC8628 similar to UP|AAR87229 (AAR87229) Putaive subtilisin-like... 94 6e-20
AV418734 91 7e-19
>TC8167 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase -
Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (65%)
Length = 2760
Score = 734 bits (1896), Expect = 0.0
Identities = 402/784 (51%), Positives = 506/784 (64%), Gaps = 20/784 (2%)
Frame = +3
Query: 9 LQSALLLLLIFCSSYTIAEKKTAQQAKKTYIIHMDKSTMPATFNDHQHWFDSSLQSVSES 68
L +LLL+ C S + KKTYI+HM+ T P + + W+ +SL+S+S +
Sbjct: 81 LSFTFILLLLQCLS------ASCSSPKKTYIVHMNHHTKPQIYPTRRDWYTASLRSLSLT 242
Query: 69 AE-------ILYTYKHVAHGFSTRLTVQEAETLAEQPGVLSVSPEVRYELHTTRTPEFLG 121
+ +LY Y +GF+ L Q+A+TL VL + + Y LHTTRTP+FLG
Sbjct: 243 TDSETSDDPLLYAYDTAYNGFAASLDEQQAQTLLGSDSVLGLYEDTLYHLHTTRTPQFLG 422
Query: 122 LLKKT------TTLSPGSDKQSQ-VVIGVLDTGVWPELKSLDDTGLSPVPSTWKGQCEAG 174
L +T TL D+ S+ V+IGVLDTGVWPE S +D G+ +PS W+G+CE
Sbjct: 423 LDTQTGLWEGHRTLE--LDQASRDVIIGVLDTGVWPESPSFNDAGMPEIPSRWRGECENA 596
Query: 175 NNMNSSSCNRKLIGARFFSKGYEATLGP---IDVSTESRSARDDDGHGSHTLTTAAGSAV 231
+ +SS CNRKLIGAR FS+G+ G E S RD DGHG+HT +TAAGS V
Sbjct: 597 TDFSSSLCNRKLIGARSFSRGFHMAAGNDGGFGKEREPPSPRDSDGHGTHTASTAAGSHV 776
Query: 232 AGASLFGLASGTARGMATQARVAAYKVCWLGGCFSSDIAAGIDKAIEDGVNIISMSIGGS 291
ASL G ASGTARGMA QARVA YKVCW GCF+SDI AG+D+AI DGV+++S+S+GG
Sbjct: 777 GNASLLGYASGTARGMAPQARVATYKVCWSDGCFASDILAGMDRAIRDGVDVLSLSLGGG 956
Query: 292 SADYFRDIIAIGAFTANSHGILVSTSAGNGGPSPSSLSNTAPWITTVGAGTIDRDFPAYI 351
S YFRD IAIGAF A GI VS SAGN GPS +SL+N APW+ TVGAGT+DRDFPA
Sbjct: 957 SGPYFRDTIAIGAFAAMERGIFVSCSAGNSGPSKASLANVAPWLMTVGAGTLDRDFPASA 1136
Query: 352 TLGNNITHTGASLYRGKPLSDSPLPLVYAGNASNFSVGYLCLPDSLVPSKVLGKIVICER 411
LGN G SLY GK + P+ LVY+ SN S G LCLP SL P+ V GK+V+C+R
Sbjct: 1137LLGNKKRFAGVSLYSGKGMGAEPVGLVYS-KGSNQS-GILCLPGSLDPAVVRGKVVLCDR 1310
Query: 412 GGNARVEKGLVVKRAGGIGMILANNEEFGEELVADSHLLPAAALGERSSKALKDYVFSSR 471
G NARVEKG VVK AGGIGMIL N GEELVADSHLLPA A+G +++YV S
Sbjct: 1311GLNARVEKGKVVKEAGGIGMILTNTAANGEELVADSHLLPAVAVGRIVGDQIREYVTSDP 1490
Query: 472 NPTAKLVFGGTHLQVKPSPVVAAFSSRGPNGLTPKILKPDLIAPGVNILAGWTGAIGPTG 531
NPTA L F GT L V+PSPVVAAFSSRGPN +T +ILKPD+I PGVNILAGW+ AIGP+G
Sbjct: 1491NPTAVLSFSGTVLNVRPSPVVAAFSSRGPNMITKQILKPDVIGPGVNILAGWSEAIGPSG 1670
Query: 532 LPVDTRHVSFNIISGTSMSCPHVSGLAAILKGSHPEWSPAAIRSALMTTSYTAYKNGQTI 591
LP D+R FNI+SGTSMSCPH+SGL A+LK +HP+WSP+AI+SALMTT+Y +
Sbjct: 1671LPQDSRKSQFNIMSGTSMSCPHISGLGALLKAAHPDWSPSAIKSALMTTAYVHDNTNSPL 1850
Query: 592 QDVATGKPATPLDFGAGHVDPVASLDPGLVYDANVDDYLGFLCALNYTSLEIKLASRRDF 651
+D A G+ +TP GAGHV+P +L PGLVYDA DY+ FLC+L+Y+ ++L +R
Sbjct: 1851RDAAGGEFSTPWAHGAGHVNPQKALSPGLVYDAKARDYIAFLCSLDYSPDHLQLIVKRAG 2030
Query: 652 KCDPKKKYRVEDFNYPSFAVPLETASGIGGGSHAPITVKYSRTLTNVGTPGTYKASVSSQ 711
+K NYPSF+V ++ A V+Y+RT+TNVG G+ V
Sbjct: 2031VNCSRKLSDPGQLNYPSFSVVFDSVV-----FDAKKVVRYTRTVTNVGEAGSVYDVVVDG 2195
Query: 712 SPSVKIAVEPQILRFQELYEKKSYTVTFTSNSMPSG---TKSFAYLYWSDGKHRVASPIA 768
V I V P L F ++ E+K YTVTF S S +F + W + +H+V SP+A
Sbjct: 2196PSMVGITVYPTKLEFGKVGERKRYTVTFVSKKGASDDLVRSAFGSITWKNEQHQVRSPVA 2375
Query: 769 ITWT 772
WT
Sbjct: 2376FAWT 2387
>CN824981
Length = 726
Score = 487 bits (1253), Expect = e-138
Identities = 241/241 (100%), Positives = 241/241 (100%)
Frame = +1
Query: 147 GVWPELKSLDDTGLSPVPSTWKGQCEAGNNMNSSSCNRKLIGARFFSKGYEATLGPIDVS 206
GVWPELKSLDDTGLSPVPSTWKGQCEAGNNMNSSSCNRKLIGARFFSKGYEATLGPIDVS
Sbjct: 4 GVWPELKSLDDTGLSPVPSTWKGQCEAGNNMNSSSCNRKLIGARFFSKGYEATLGPIDVS 183
Query: 207 TESRSARDDDGHGSHTLTTAAGSAVAGASLFGLASGTARGMATQARVAAYKVCWLGGCFS 266
TESRSARDDDGHGSHTLTTAAGSAVAGASLFGLASGTARGMATQARVAAYKVCWLGGCFS
Sbjct: 184 TESRSARDDDGHGSHTLTTAAGSAVAGASLFGLASGTARGMATQARVAAYKVCWLGGCFS 363
Query: 267 SDIAAGIDKAIEDGVNIISMSIGGSSADYFRDIIAIGAFTANSHGILVSTSAGNGGPSPS 326
SDIAAGIDKAIEDGVNIISMSIGGSSADYFRDIIAIGAFTANSHGILVSTSAGNGGPSPS
Sbjct: 364 SDIAAGIDKAIEDGVNIISMSIGGSSADYFRDIIAIGAFTANSHGILVSTSAGNGGPSPS 543
Query: 327 SLSNTAPWITTVGAGTIDRDFPAYITLGNNITHTGASLYRGKPLSDSPLPLVYAGNASNF 386
SLSNTAPWITTVGAGTIDRDFPAYITLGNNITHTGASLYRGKPLSDSPLPLVYAGNASNF
Sbjct: 544 SLSNTAPWITTVGAGTIDRDFPAYITLGNNITHTGASLYRGKPLSDSPLPLVYAGNASNF 723
Query: 387 S 387
S
Sbjct: 724 S 726
>TC16670 similar to PIR|JC7519|JC7519 subtilisin-like serine proteinase -
Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (42%)
Length = 1183
Score = 451 bits (1159), Expect = e-127
Identities = 222/341 (65%), Positives = 264/341 (77%)
Frame = +2
Query: 15 LLLIFCSSYTIAEKKTAQQAKKTYIIHMDKSTMPATFNDHQHWFDSSLQSVSESAEILYT 74
LL++F S+ I + K TYI+H+ KS MP +F H W++SSLQSVS+SAE+LYT
Sbjct: 173 LLVLFLSTLCI---HATPENKGTYIVHVAKSEMPESFEHHAAWYESSLQSVSDSAEMLYT 343
Query: 75 YKHVAHGFSTRLTVQEAETLAEQPGVLSVSPEVRYELHTTRTPEFLGLLKKTTTLSPGSD 134
Y + HGFSTRLT +EA L Q GVL+V PE ++ELHTTRTP FLGL K+ + P S
Sbjct: 344 YDNAIHGFSTRLTPEEARLLEAQTGVLAVLPERKFELHTTRTPLFLGL-DKSADMFPASG 520
Query: 135 KQSQVVIGVLDTGVWPELKSLDDTGLSPVPSTWKGQCEAGNNMNSSSCNRKLIGARFFSK 194
+V++GVLDTGVWPE KS DDTG PVPSTWKG+CE+G N +++CN+KLIGAR+FSK
Sbjct: 521 AGGEVIVGVLDTGVWPESKSFDDTGFGPVPSTWKGECESGTNFTAANCNKKLIGARYFSK 700
Query: 195 GYEATLGPIDVSTESRSARDDDGHGSHTLTTAAGSAVAGASLFGLASGTARGMATQARVA 254
G EA LGPID + ES+S RDDDGHG+HT +TAAGS V+GASLFG A+GTARGMA ARVA
Sbjct: 701 GAEAMLGPIDETAESKSPRDDDGHGTHTASTAAGSVVSGASLFGYAAGTARGMAPHARVA 880
Query: 255 AYKVCWLGGCFSSDIAAGIDKAIEDGVNIISMSIGGSSADYFRDIIAIGAFTANSHGILV 314
AYKVCW GGCFSSDI A IDKAI D VN++S+S+GG ADY+RD +AIGAF A GILV
Sbjct: 881 AYKVCWKGGCFSSDILAAIDKAIADNVNVLSLSLGGGMADYYRDSVAIGAFAAMEKGILV 1060
Query: 315 STSAGNGGPSPSSLSNTAPWITTVGAGTIDRDFPAYITLGN 355
S SAGN GPS SLSN APWITTVGAGT+DRDFPA + LGN
Sbjct: 1061SCSAGNAGPSAYSLSNVAPWITTVGAGTLDRDFPALVELGN 1183
>TC10429 similar to UP|Q9ZR46 (Q9ZR46) P69C protein, partial (52%)
Length = 1742
Score = 411 bits (1057), Expect = e-115
Identities = 245/559 (43%), Positives = 328/559 (57%), Gaps = 8/559 (1%)
Frame = +3
Query: 24 TIAEKKTAQQAKKTYIIHMD--KSTMPATFNDHQHWFDS----SLQSVSESAEILYTYKH 77
T + T + K YIIH+ + M D + W+ S +L+S E ++Y+YK+
Sbjct: 111 TTTTESTETSSSKIYIIHVTGPEGKMLTESEDLESWYHSFLPPTLKSSEEQPRVIYSYKN 290
Query: 78 VAHGFSTRLTVQEAETLAEQPGVLSVSPEVRYELHTTRTPEFLGLLKKTTTLSPGSDKQS 137
V GF+ LT +E + + + + Y + P+FLGL ++ T + S+
Sbjct: 291 VLRGFAASLTQEELSAVERKMALFQLILRGCYIVKPLIPPKFLGL-QQDTGVWKESNFGK 467
Query: 138 QVVIGVLDTGVWPELKSLDDTGLSPVPSTWKGQCEAGNNMNSSSCNRKLIGARFFSKGYE 197
V+IGVLD+G+ P S D G+ P P WKG+C+ +N ++CN KLIGAR F+ E
Sbjct: 468 GVIIGVLDSGITPGHPSFSDVGIPPPPPKWKGRCD----LNVTACNNKLIGARAFNLAAE 635
Query: 198 ATLGPIDVSTESRSARDDDGHGSHTLTTAAGSAVAGASLFGLASGTARGMATQARVAAYK 257
A G ++ + D+DGHG+HT +TAAG+ V A + G A GTA GMA A +A YK
Sbjct: 636 AMNGK-----KAEAPIDEDGHGTHTASTAAGAFVNYAEVLGNAKGTAAGMAPHAHLAIYK 800
Query: 258 VCWLGGCFSSDIAAGIDKAIEDGVNIISMSIGGSSAD-YFRDIIAIGAFTANSHGILVST 316
VC+ C SDI A +D A+EDGV++IS+S+G S +F D AIGAF A GI VS
Sbjct: 801 VCFGEDCPESDILAALDAAVEDGVDVISISLGLSEPPPFFNDSTAIGAFAAMQKGIFVSC 980
Query: 317 SAGNGGPSPSSLSNTAPWITTVGAGTIDRDFPAYITLGNNITHTGASLYRGKPLSDSPLP 376
+AGN GP SS+ N APWI TVGA TIDR A LGN G S+++ + + LP
Sbjct: 981 AAGNSGPFNSSIVNAAPWILTVGASTIDRRIVATAKLGNGQEFDGESVFQPSSFTPTLLP 1160
Query: 377 LVYAGNASNFSVGYLCLPDSLVPSKVLGKIVICERGGN-ARVEKGLVVKRAGGIGMILAN 435
L YAG + C SL S GK+V+CERGG AR+ KG VKRAGG MIL N
Sbjct: 1161LAYAGKNGKEESAF-CANGSLDDSAFRGKVVLCERGGGIARIAKGEEVKRAGGAAMILMN 1337
Query: 436 NEEFGEELVADSHLLPAAALGERSSKALKDYVFSSRNPTAKLVFGGTHLQVKPSPVVAAF 495
+E L AD H LPA + + +K Y+ S+ PTA ++F GT + +P VA+F
Sbjct: 1338DETNAFSLSADVHALPATHVSYAAGIEIKAYINSTATPTATILFKGTVIGNSLAPAVASF 1517
Query: 496 SSRGPNGLTPKILKPDLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNIISGTSMSCPHVS 555
SSRGPN +P ILKPD+I PGVNILA W P D++ ++FNI SGTSMSCPH+S
Sbjct: 1518SSRGPNLPSPGILKPDIIGPGVNILAAWP---FPLSNSTDSK-LTFNIESGTSMSCPHLS 1685
Query: 556 GLAAILKGSHPEWSPAAIR 574
G+AA+LK SHP WSPAAI+
Sbjct: 1686GIAALLKSSHPHWSPAAIK 1742
>TC13087 similar to UP|Q9FIF8 (Q9FIF8) Serine protease-like protein, partial
(31%)
Length = 1036
Score = 241 bits (616), Expect = 2e-64
Identities = 142/352 (40%), Positives = 200/352 (56%), Gaps = 2/352 (0%)
Frame = +2
Query: 206 STESRSARDDDGHGSHTLTTAAGSAVAGASLFGLASGTARGMATQARVAAYKVCWLG-GC 264
S + RD +GHGSHT +TAAG+ V GAS +GLA GTARG AR+AAYKVC+ GC
Sbjct: 5 SARGNTVRDIEGHGSHTASTAAGNTVIGASFYGLAQGTARGAVPSARIAAYKVCYPNHGC 184
Query: 265 FSSDIAAGIDKAIEDGVNIISMSIGGS-SADYFRDIIAIGAFTANSHGILVSTSAGNGGP 323
++I A D AI DGVNI+S+S+G + D D +AIG+F A GIL +AGN GP
Sbjct: 185 DDANILAAFDDAIADGVNILSVSLGTNYCVDLLSDTVAIGSFHAMERGILTLQAAGNSGP 364
Query: 324 SPSSLSNTAPWITTVGAGTIDRDFPAYITLGNNITHTGASLYRGKPLSDSPLPLVYAGNA 383
+ S+ + APW+ TV A TIDR F + LGN +T G S+ + + P+ +
Sbjct: 365 ASPSICSVAPWLLTVAASTIDRQFIDKVLLGNGMTLIGRSV-NAVVSNGTKFPIAKWNAS 541
Query: 384 SNFSVGYLCLPDSLVPSKVLGKIVICERGGNARVEKGLVVKRAGGIGMILANNEEFGEEL 443
G+ L L V GK+V+CE G E + G I L N +
Sbjct: 542 DERCKGFSDLCQCLGSESVKGKVVLCE--GRMHDEAAYLDGAVGSITEDLTNPDND---- 703
Query: 444 VADSHLLPAAALGERSSKALKDYVFSSRNPTAKLVFGGTHLQVKPSPVVAAFSSRGPNGL 503
V+ P+ L + +++Y S++NP A+++ + + +P V +FSSRGPN L
Sbjct: 704 VSFVTYFPSVNLNPKDYAIVQNYTNSAKNPKAEILKSEI-FKDRSAPKVVSFSSRGPNPL 880
Query: 504 TPKILKPDLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNIISGTSMSCPHVS 555
P+I+KPD+ APGV+ILA W+ P+ D R V +N++SGTSM+CPHV+
Sbjct: 881 IPEIMKPDVSAPGVDILAAWSPEAAPSEYSSDKRKVRYNVVSGTSMACPHVA 1036
>AV426475
Length = 300
Score = 201 bits (512), Expect = 3e-52
Identities = 98/99 (98%), Positives = 99/99 (99%)
Frame = +2
Query: 464 KDYVFSSRNPTAKLVFGGTHLQVKPSPVVAAFSSRGPNGLTPKILKPDLIAPGVNILAGW 523
+DYVFSSRNPTAKLVFGGTHLQVKPSPVVAAFSSRGPNGLTPKILKPDLIAPGVNILAGW
Sbjct: 2 RDYVFSSRNPTAKLVFGGTHLQVKPSPVVAAFSSRGPNGLTPKILKPDLIAPGVNILAGW 181
Query: 524 TGAIGPTGLPVDTRHVSFNIISGTSMSCPHVSGLAAILK 562
TGAIGPTGLPVDTRHVSFNIISGTSMSCPHVSGLAAILK
Sbjct: 182 TGAIGPTGLPVDTRHVSFNIISGTSMSCPHVSGLAAILK 298
>AU088794
Length = 433
Score = 187 bits (474), Expect = 7e-48
Identities = 85/144 (59%), Positives = 108/144 (74%)
Frame = +2
Query: 491 VVAAFSSRGPNGLTPKILKPDLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNIISGTSMS 550
VVA+FS+RGPN +P+ILKPD+I PG+NILA W GP+G+P D R FNI+SGTSM+
Sbjct: 2 VVASFSARGPNPESPEILKPDVIXPGLNILAAWPDXXGPSGVPSDVRRTEFNILSGTSMA 181
Query: 551 CPHVSGLAAILKGSHPEWSPAAIRSALMTTSYTAYKNGQTIQDVATGKPATPLDFGAGHV 610
CPHVSGLAA+LK +HP WSPAAI+SALMTT+YT G + D + G + D+G+GHV
Sbjct: 182 CPHVSGLAALLKAAHPXWSPAAIKSALMTTAYTVDNKGDAMLDESNGNVSLVFDYGSGHV 361
Query: 611 DPVASLDPGLVYDANVDDYLGFLC 634
P ++DPGLVYD + DY+ FLC
Sbjct: 362 HPXKAMDPGLVYDISTYDYVDFLC 433
>AV408408
Length = 429
Score = 185 bits (469), Expect = 3e-47
Identities = 89/141 (63%), Positives = 113/141 (80%)
Frame = +3
Query: 384 SNFSVGYLCLPDSLVPSKVLGKIVICERGGNARVEKGLVVKRAGGIGMILANNEEFGEEL 443
SN + G LC+ +L P KV GKIV+C+RG NARV+KG VVK AGG+GM+L+N GEEL
Sbjct: 6 SNTTNGNLCMMGTLSPDKVAGKIVMCDRGMNARVQKGQVVKGAGGLGMVLSNTAANGEEL 185
Query: 444 VADSHLLPAAALGERSSKALKDYVFSSRNPTAKLVFGGTHLQVKPSPVVAAFSSRGPNGL 503
VAD+HLLP +A+GE++ A+K Y+FS T K++F GT + ++PSPVVAAFSSRGPN +
Sbjct: 186 VADAHLLPTSAVGEKAGNAIKKYLFSDPKATVKILFEGTKVGIQPSPVVAAFSSRGPNSI 365
Query: 504 TPKILKPDLIAPGVNILAGWT 524
TP+ILKPDLIAPGVNILAGW+
Sbjct: 366 TPQILKPDLIAPGVNILAGWS 428
>TC17789 similar to PIR|T05768|T05768 subtilisin-like proteinase -
Arabidopsis thaliana {Arabidopsis
thaliana;} , partial (14%)
Length = 940
Score = 182 bits (461), Expect = 2e-46
Identities = 101/228 (44%), Positives = 133/228 (58%), Gaps = 6/228 (2%)
Frame = +2
Query: 549 MSCPHVSGLAAILKGSHPEWSPAAIRSALMTTSYTAYKNGQTIQDVATGKPATPLDFGAG 608
M+CPH+SG AA+LK +HP+WSPAA+RSA+MTT+ Q + D ATG +TP DFGAG
Sbjct: 2 MACPHISGAAALLKSAHPDWSPAAVRSAMMTTATVLDNRKQILTDEATGNSSTPYDFGAG 181
Query: 609 HVDPVASLDPGLVYDANVDDYLGFLCALNYTSLEIKLASRRDFKCDPKKKYRVEDFNYPS 668
HV+ ++DPGLVYD DY+ FLC + Y I++ +R C P +K E+ NYPS
Sbjct: 182 HVNLGLAMDPGLVYDITGADYVNFLCGIGYGPRVIQVITRAPVSC-PARKPSPENLNYPS 358
Query: 669 FAVPLETASGIGGGSHAPITVKYSRTLTNVGTPGT-YKASVSSQSPSVKIAVEPQILRFQ 727
F GS T + RT+TNVG + Y+ SV SQ V + V+P L F
Sbjct: 359 FVAMFPV------GSKGLTTKTFIRTVTNVGPANSVYRVSVESQMKGVTVTVKPSRLVFS 520
Query: 728 ELYEKKSYTVTFTSNSM-----PSGTKSFAYLYWSDGKHRVASPIAIT 770
E +K+SY VT + + PSG F L W+DGKH V SPI +T
Sbjct: 521 EAVKKRSYVVTVAAETRFLKMGPSGA-VFGSLTWTDGKHVVRSPIVVT 661
>TC13765 similar to UP|Q9LLL8 (Q9LLL8) Subtilisin-type serine endopeptidase
XSP1, partial (20%)
Length = 635
Score = 152 bits (384), Expect = 2e-37
Identities = 86/190 (45%), Positives = 126/190 (66%)
Frame = +1
Query: 392 CLPDSLVPSKVLGKIVICERGGNARVEKGLVVKRAGGIGMILANNEEFGEELVADSHLLP 451
C DSL P+KV GK+V C+ G N E VVK+ GGIG I+ ++++ + +A + P
Sbjct: 79 CYEDSLQPNKVKGKLVYCKLG-NWGTEG--VVKKFGGIGSIM-ESDQYPD--LAQIFMAP 240
Query: 452 AAALGERSSKALKDYVFSSRNPTAKLVFGGTHLQVKPSPVVAAFSSRGPNGLTPKILKPD 511
A L +++ +Y+ S+R+P+A V TH + P+P VA FSSRGPN + +LKPD
Sbjct: 241 ATILNHTIGESVTNYIKSTRSPSA--VIYKTHEEKCPAPFVATFSSRGPNPGSHNVLKPD 414
Query: 512 LIAPGVNILAGWTGAIGPTGLPVDTRHVSFNIISGTSMSCPHVSGLAAILKGSHPEWSPA 571
+ APG++ILA +T TG DT+ F+++SGTSM+CPHV+G+AA +K HP W+PA
Sbjct: 415 IAAPGIDILASYTLRKSITGSEGDTQFSEFSLLSGTSMACPHVAGVAAYVKSFHPNWTPA 594
Query: 572 AIRSALMTTS 581
AIRSA++TT+
Sbjct: 595 AIRSAIITTA 624
>AV421923
Length = 463
Score = 148 bits (374), Expect = 3e-36
Identities = 77/153 (50%), Positives = 98/153 (63%), Gaps = 11/153 (7%)
Frame = +2
Query: 471 RNPTAKLVFGGTHLQVKPSPVVAAFSSRGPNGLTPKILKPDLIAPGVNILAGWTGAIGPT 530
+ PTA + F GT P+PVVAAFSSRGP+ + ++KPD+ APGV+ILA W P+
Sbjct: 2 KKPTASISFIGTRFG-DPAPVVAAFSSRGPSIVGLDVIKPDVTAPGVHILAAWPSKTSPS 178
Query: 531 GLPVDTRHVSFNIISGTSMSCPHVSGLAAILKGSHPEWSPAAIRSALMTTSYTAYKNGQT 590
L D R V FNIISGTSMSCPHVSG+AA+++ H +WSPAA++SALMTT+YT G
Sbjct: 179 MLKNDKRRVLFNIISGTSMSCPHVSGVAALIRSVHRDWSPAAVKSALMTTAYTLNNKGAP 358
Query: 591 IQDVATGKP-----------ATPLDFGAGHVDP 612
I D+ A P FG+GHV+P
Sbjct: 359 ISDIGASNSISNSNSNHSGFAVPFAFGSGHVNP 457
>AV420480
Length = 412
Score = 145 bits (367), Expect = 2e-35
Identities = 72/138 (52%), Positives = 97/138 (70%)
Frame = +3
Query: 480 GGTHLQVKPSPVVAAFSSRGPNGLTPKILKPDLIAPGVNILAGWTGAIGPTGLPVDTRHV 539
GGT + +P VA FS+RGP+ P ILKPD++APGVNI+A W +GPT LP D R V
Sbjct: 3 GGTVIGNSRAPAVATFSARGPSFTNPSILKPDVVAPGVNIIAAWPQNLGPTSLPQDLRRV 182
Query: 540 SFNIISGTSMSCPHVSGLAAILKGSHPEWSPAAIRSALMTTSYTAYKNGQTIQDVATGKP 599
+F+++SGTSMSCPHVSG+AA++ +HP+WSPAAI+SA+MTT+ + I D KP
Sbjct: 183 NFSVMSGTSMSCPHVSGIAALVHSAHPKWSPAAIKSAIMTTADVTDHMKRPILD--EDKP 356
Query: 600 ATPLDFGAGHVDPVASLD 617
A GAG+V+P +L+
Sbjct: 357 AGVFAIGAGNVNPQRALN 410
>TC11984 similar to UP|Q93WQ0 (Q93WQ0) Subtilisin-type protease, partial
(30%)
Length = 966
Score = 144 bits (362), Expect = 7e-35
Identities = 95/244 (38%), Positives = 125/244 (50%), Gaps = 6/244 (2%)
Frame = +1
Query: 532 LPVDTRHVSFNIISGTSMSCPHVSGLAAILKGSHPEWSPAAIRSALMTTSYTAYKNGQTI 591
+P + +NIISGTSMSCPHVSGLA +K H WS +AI+SA+M TS T N +
Sbjct: 31 VPKGKKPSQYNIISGTSMSCPHVSGLAGSIKSRHTTWSASAIKSAIM-TSATQNNNLKAP 207
Query: 592 QDVATGKPATPLDFGAGHVDPVASLDPGLVYDANVDDYLGFLCALNYTSLEIKLASRR-- 649
+G ATP D+GAG + AS PGLVY+ + DYL +LC + IK+ SR
Sbjct: 208 MTTDSGSVATPYDYGAGEMTTSASFQPGLVYETSTTDYLNYLCYIGLNVTTIKVISRTVP 387
Query: 650 -DFKC-DPKKKYRVEDFNYPSFAVPLETASGIGGGSHAPITVKYSRTLTNVGTPGTYKAS 707
F C V + NYPS A+ G + SRT+TNVG S
Sbjct: 388 DSFTCPQDSSADHVSNINYPSIAITNFNGKG---------AINVSRTVTNVGEEDETVYS 540
Query: 708 VSSQSPS-VKIAVEPQILRFQELYEKKSYTVTFTSNSMPSGTKS-FAYLYWSDGKHRVAS 765
+PS V I + P+ L F + +K SY V F+ S S + F L WS+GK+ V S
Sbjct: 541 PIVDAPSGVNIKLIPEKLPFTKSSQKLSYPVIFSLTSSTSLKEDLFGSLAWSNGKYIVRS 720
Query: 766 PIAI 769
P +
Sbjct: 721 PFVL 732
>TC19089 similar to UP|Q9ZTT3 (Q9ZTT3) Subtilisin-like protease C1, partial
(16%)
Length = 600
Score = 131 bits (329), Expect = 5e-31
Identities = 67/155 (43%), Positives = 92/155 (59%)
Frame = +1
Query: 499 GPNGLTPKILKPDLIAPGVNILAGWTGAIGPTGLPVDTRHVSFNIISGTSMSCPHVSGLA 558
GPN +TP ILKPD+ APGV +LA W+ + + D R + +N ISGTSM+CPH + A
Sbjct: 16 GPNPITPNILKPDIAAPGVTVLAAWSPLNPISEVEGDKRKLPYNAISGTSMACPHAAAAA 195
Query: 559 AILKGSHPEWSPAAIRSALMTTSYTAYKNGQTIQDVATGKPATPLDFGAGHVDPVASLDP 618
A +K HP WSPA I+SALMTT+ T + A P +GAG ++PV +++P
Sbjct: 196 AYVKSFHPNWSPAMIKSALMTTA--------TPMNSAL-NPEAEFAYGAGQINPVKAVNP 348
Query: 619 GLVYDANVDDYLGFLCALNYTSLEIKLASRRDFKC 653
GLVYD DY+ FLC YT ++ ++ C
Sbjct: 349 GLVYDITEQDYIQFLCGXGYTDTLLQNLTQHKRXC 453
>TC19209 similar to GB|BAB11244.1|10177874|AB010074 serine protease-like
protein {Arabidopsis thaliana;} , partial (12%)
Length = 553
Score = 129 bits (325), Expect = 1e-30
Identities = 75/181 (41%), Positives = 103/181 (56%), Gaps = 16/181 (8%)
Frame = +1
Query: 1 MKMPIFQLLQSALLLLLIFCSSYTIAEKKTAQQAKKTYIIHMDKSTMPA-TFNDHQHWFD 59
M + F ++ +L + +I AQ KKTYII +D S MPA TF+ H W+
Sbjct: 10 MTLSSFTPMEKMSCILAKYLLFLSITLSANAQFLKKTYIIQIDNSAMPAGTFSTHLEWYS 189
Query: 60 SSLQSV-------------SESAEILYTYKHVAHGFSTRLTVQEAETLAEQPGVLSVSPE 106
S ++SV E I+Y+Y HG + +LT +EA+ L Q GV+++ PE
Sbjct: 190 SKVKSVLSKSLESETISSEEEEERIIYSYHTAFHGVAAKLTPEEAQKLESQEGVVAIFPE 369
Query: 107 VRYELHTTRTPEFLGLLKKTTTLSPGSDKQS--QVVIGVLDTGVWPELKSLDDTGLSPVP 164
RY+LHTTR+P FLGL K +T S+K S VV+GV DTG+WPE +S +DTG PVP
Sbjct: 370 TRYQLHTTRSPTFLGLEKMQST-EIWSEKLSNHDVVVGVSDTGIWPESQSFNDTGFKPVP 546
Query: 165 S 165
S
Sbjct: 547 S 549
>TC16297 weakly similar to UP|Q93WQ0 (Q93WQ0) Subtilisin-type protease,
partial (10%)
Length = 574
Score = 123 bits (308), Expect = 1e-28
Identities = 76/176 (43%), Positives = 106/176 (60%), Gaps = 10/176 (5%)
Frame = +2
Query: 268 DIAAGIDKAIEDGVNIISMSIGG----SSADYFRDIIAIGAFTANSHGILVSTSAGNGGP 323
D+ A ID+AI DGV++ S+S GG S+ + F D ++IG+ A + ILV SAGN GP
Sbjct: 2 DVLAAIDQAITDGVDVNSVSAGGRSIISAEEIFTDEVSIGSVHALAKNILVVASAGNDGP 181
Query: 324 SPSSLSNTAPWITTVGAGTIDRDFPAYITLGNNITHTGASLYRGKPLSDSPLPLVYA--- 380
+ ++ N APW+ TV A TIDR+F + ITLGNN TGASL+ P + L+ +
Sbjct: 182 ALGTVLNVAPWVFTVAASTIDREFSSTITLGNNQQITGASLFVNLP-PNQLFSLILSTDA 358
Query: 381 --GNASNFSVGYLCLPDSLVPSKVLGKIVICERGGNAR-VEKGLVVKRAGGIGMIL 433
NA+N LC P +L P+KV GK+V+C RGG + V +G +G GM+L
Sbjct: 359 KLANATNRD-AQLCRPGTLDPAKVKGKVVVCTRGGKIKSVAEGSEALSSGAKGMVL 523
>TC8535 weakly similar to UP|Q9FJF3 (Q9FJF3) Serine protease-like protein,
partial (10%)
Length = 780
Score = 108 bits (269), Expect = 4e-24
Identities = 65/171 (38%), Positives = 93/171 (54%), Gaps = 2/171 (1%)
Frame = +2
Query: 601 TPLDFGAGHVDPVASLDPGLVYDANVDDYLGFLCALNYTSLEIK-LASRRDFKCDPKKKY 659
TP +G+GHV P +++DPGLVYD ++ DYL FLCA Y I L + F C +
Sbjct: 2 TPFAYGSGHVQPNSAIDPGLVYDLSIVDYLNFLCASGYDQQLISALNFNKTFTC--SGSH 175
Query: 660 RVEDFNYPSFAVPLETASGIGGGSHAPITVKYSRTLTNVGTPGTYKASVSSQSPSVKIAV 719
+ D NYPS +P ++ V +RT+TNVG P Y A ++Q P KI V
Sbjct: 176 SITDLNYPSITLPNIRSN----------VVNVTRTVTNVGPPSKYFA--NAQLPGHKIVV 319
Query: 720 EPQILRFQELYEKKSYTVTFTSNSMPSGTK-SFAYLYWSDGKHRVASPIAI 769
P L F+++ EKK++ V + S+ K F L W++G+H V SPI +
Sbjct: 320 VPNSLSFKKIGEKKTFQVIVQATSVTQRRKYLFGKLQWTNGRHIVRSPITV 472
>TC13483 weakly similar to UP|Q9FIF8 (Q9FIF8) Serine protease-like protein,
partial (11%)
Length = 550
Score = 94.4 bits (233), Expect = 6e-20
Identities = 61/159 (38%), Positives = 88/159 (54%), Gaps = 9/159 (5%)
Frame = +3
Query: 620 LVYDANVDDYLGFLCALNYTSLEIKL-ASRRDFKC-DPKKKYRVEDFNYPSFAV---PLE 674
LVYD ++ DY+ LC Y++ ++K+ A C + V+D NYP+ V PLE
Sbjct: 3 LVYDISMKDYVNMLCNYGYSAKKLKIIAGAHSLSCYGEHHRSLVKDLNYPAITVNVKPLE 182
Query: 675 TASGIGGGSHAPITVKYSRTLTNVGTPG--TYKASVSSQSPSVK--IAVEPQILRFQELY 730
P +K +RT+TNVG P TYKA+V P++K I VEP++LRF+ L+
Sbjct: 183 -----------PYIIKVNRTVTNVGFPHNLTYKATVL---PNLKMNITVEPEVLRFESLH 320
Query: 731 EKKSYTVTFTSNSMPSGTKSFAYLYWSDGKHRVASPIAI 769
EKKS+ V + + + + L WSDGKH V SPI +
Sbjct: 321 EKKSFIVMVSGGGLRISSAISSSLVWSDGKHYVKSPITV 437
>TC8628 similar to UP|AAR87229 (AAR87229) Putaive subtilisin-like
proteinase, partial (5%)
Length = 547
Score = 94.4 bits (233), Expect = 6e-20
Identities = 47/82 (57%), Positives = 63/82 (76%), Gaps = 1/82 (1%)
Frame = +1
Query: 691 YSRTLTNVGTPGTYKASVSSQSPSVKIAVEPQILRFQELYEKKSYTVTFTSN-SMPSGTK 749
++R+L+NVG GTYKASV+ +PSVK+++EPQ+L F+E EKKSYTVTFTS+ S P
Sbjct: 1 HTRSLSNVGPAGTYKASVTKNNPSVKVSIEPQVLSFKE-SEKKSYTVTFTSSGSAPQRVN 177
Query: 750 SFAYLYWSDGKHRVASPIAITW 771
F L WSDGK+ V +PI+I+W
Sbjct: 178 GFERLEWSDGKNVVGTPISISW 243
>AV418734
Length = 287
Score = 90.9 bits (224), Expect = 7e-19
Identities = 45/99 (45%), Positives = 63/99 (63%)
Frame = +1
Query: 514 APGVNILAGWTGAIGPTGLPVDTRHVSFNIISGTSMSCPHVSGLAAILKGSHPEWSPAAI 573
APG+NILA W+ A G FNI+SGTSM+CPHV+G+A ++K HP WSP+AI
Sbjct: 10 APGLNILAAWSPAAGNM----------FNIVSGTSMACPHVTGIATLVKAVHPSWSPSAI 159
Query: 574 RSALMTTSYTAYKNGQTIQDVATGKPATPLDFGAGHVDP 612
+SA+MTT+ K+ + I + A D+G+G V+P
Sbjct: 160 KSAIMTTATILDKHHRHISADPEQRTANAFDYGSGFVNP 276
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.315 0.132 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,389,855
Number of Sequences: 28460
Number of extensions: 165107
Number of successful extensions: 963
Number of sequences better than 10.0: 71
Number of HSP's better than 10.0 without gapping: 912
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 917
length of query: 772
length of database: 4,897,600
effective HSP length: 97
effective length of query: 675
effective length of database: 2,136,980
effective search space: 1442461500
effective search space used: 1442461500
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Lotus: description of TM0096b.8


