

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0096b.1
(137 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
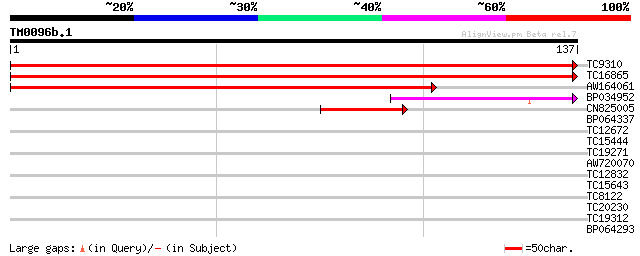
Score E
Sequences producing significant alignments: (bits) Value
TC9310 similar to UP|RS24_ARATH (Q9SS17) 40S ribosomal protein S... 271 4e-74
TC16865 similar to UP|RS24_ARATH (Q9SS17) 40S ribosomal protein ... 268 2e-73
AW164061 202 2e-53
BP034952 68 6e-13
CN825005 43 2e-05
BP064337 27 1.2
TC12672 weakly similar to UP|Q9LV59 (Q9LV59) Genomic DNA, chromo... 27 1.6
TC15444 26 2.1
TC19271 similar to UP|Q8S531 (Q8S531) Cytosolic aldehyde dehydro... 26 2.8
AW720070 25 3.6
TC12832 weakly similar to UP|POLX_TOBAC (P10978) Retrovirus-rela... 25 4.7
TC15643 similar to UP|O64720 (O64720) At2g02560 protein, partial... 25 6.2
TC8122 similar to UP|Q94A43 (Q94A43) AT4g36780/C7A10_580, partia... 24 8.1
TC20230 homologue to UP|CYC_BRAOL (P00050) Cytochrome c, complete 24 8.1
TC19312 similar to UP|Q9XIS4 (Q9XIS4) Branching enzyme 3 , part... 24 8.1
BP064293 24 8.1
>TC9310 similar to UP|RS24_ARATH (Q9SS17) 40S ribosomal protein S24,
complete
Length = 706
Score = 271 bits (692), Expect = 4e-74
Identities = 137/137 (100%), Positives = 137/137 (100%)
Frame = +1
Query: 1 MADKAVTIRTRKFMTNRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKDPNTVFVF 60
MADKAVTIRTRKFMTNRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKDPNTVFVF
Sbjct: 85 MADKAVTIRTRKFMTNRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKDPNTVFVF 264
Query: 61 KFRTHFGGGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDTKVEKSRKQLKERKNRAKKIR 120
KFRTHFGGGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDTKVEKSRKQLKERKNRAKKIR
Sbjct: 265 KFRTHFGGGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDTKVEKSRKQLKERKNRAKKIR 444
Query: 121 GVKKTKAADAAKAGKKK 137
GVKKTKAADAAKAGKKK
Sbjct: 445 GVKKTKAADAAKAGKKK 495
>TC16865 similar to UP|RS24_ARATH (Q9SS17) 40S ribosomal protein S24,
complete
Length = 634
Score = 268 bits (685), Expect = 2e-73
Identities = 135/137 (98%), Positives = 136/137 (98%)
Frame = +2
Query: 1 MADKAVTIRTRKFMTNRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKDPNTVFVF 60
MADKAVTIRTRKFMTNRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKDPNTVFVF
Sbjct: 68 MADKAVTIRTRKFMTNRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKDPNTVFVF 247
Query: 61 KFRTHFGGGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDTKVEKSRKQLKERKNRAKKIR 120
KFRTHFGGGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDTKVEKSRKQLKERKNRAKKIR
Sbjct: 248 KFRTHFGGGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDTKVEKSRKQLKERKNRAKKIR 427
Query: 121 GVKKTKAADAAKAGKKK 137
GVKKTKA+DAAK GKKK
Sbjct: 428 GVKKTKASDAAKGGKKK 478
>AW164061
Length = 340
Score = 202 bits (513), Expect = 2e-53
Identities = 100/103 (97%), Positives = 101/103 (97%)
Frame = +1
Query: 1 MADKAVTIRTRKFMTNRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKDPNTVFVF 60
MADKAVTIRTRKFMTNRLLSRKQFIVDVLHPGRANVSK ELKEKLARLYDVKDPNTVFVF
Sbjct: 31 MADKAVTIRTRKFMTNRLLSRKQFIVDVLHPGRANVSKTELKEKLARLYDVKDPNTVFVF 210
Query: 61 KFRTHFGGGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDTKVE 103
KFRTHFGGGKSTGFGLIY SLENA+KYEPKYRLIRNGLDTKVE
Sbjct: 211 KFRTHFGGGKSTGFGLIYHSLENAQKYEPKYRLIRNGLDTKVE 339
>BP034952
Length = 450
Score = 67.8 bits (164), Expect = 6e-13
Identities = 41/73 (56%), Positives = 43/73 (58%), Gaps = 28/73 (38%)
Frame = -3
Query: 93 LIRNGLDTKVEKSRKQLKERKNRAKKIRGVKK---------------------------- 124
L +NGLDTKVEKSRKQLKERKNRAKKIRGVKK
Sbjct: 334 LFQNGLDTKVEKSRKQLKERKNRAKKIRGVKKVIKAYNS*WFLTLYLGFVELNWDVLFLQ 155
Query: 125 TKAADAAKAGKKK 137
TKA+DAAK GKKK
Sbjct: 154 TKASDAAKGGKKK 116
>CN825005
Length = 665
Score = 42.7 bits (99), Expect = 2e-05
Identities = 19/21 (90%), Positives = 19/21 (90%)
Frame = +2
Query: 76 LIYDSLENAKKYEPKYRLIRN 96
LIYDSLENAKKYEPKYR I N
Sbjct: 599 LIYDSLENAKKYEPKYRFIMN 661
>BP064337
Length = 485
Score = 26.9 bits (58), Expect = 1.2
Identities = 11/22 (50%), Positives = 14/22 (63%)
Frame = -2
Query: 46 ARLYDVKDPNTVFVFKFRTHFG 67
AR+Y V TVF FK + +FG
Sbjct: 160 ARMYQVNGDETVFFFKAKDYFG 95
>TC12672 weakly similar to UP|Q9LV59 (Q9LV59) Genomic DNA, chromosome 3, P1
clone: MOB24, partial (14%)
Length = 548
Score = 26.6 bits (57), Expect = 1.6
Identities = 13/40 (32%), Positives = 24/40 (59%), Gaps = 1/40 (2%)
Frame = +3
Query: 74 FGLIYDSLENAKKYE-PKYRLIRNGLDTKVEKSRKQLKER 112
FG IY+ +EN K+ + + +R + ++E +KQ+ ER
Sbjct: 3 FGKIYEKIENTKRQQMMELEKMRLDFNRELELQKKQILER 122
>TC15444
Length = 641
Score = 26.2 bits (56), Expect = 2.1
Identities = 12/19 (63%), Positives = 14/19 (73%)
Frame = +2
Query: 20 SRKQFIVDVLHPGRANVSK 38
+R +VDVLH GRAN SK
Sbjct: 86 NRVLHVVDVLHRGRANKSK 142
>TC19271 similar to UP|Q8S531 (Q8S531) Cytosolic aldehyde dehydrogenase
RF2C, partial (25%)
Length = 485
Score = 25.8 bits (55), Expect = 2.8
Identities = 15/33 (45%), Positives = 16/33 (48%)
Frame = +1
Query: 55 NTVFVFKFRTHFGGGKSTGFGLIYDSLENAKKY 87
N F F FGG K +GFG Y LE KY
Sbjct: 229 NCFFAFDIDCPFGGYKMSGFGRDY-GLEALHKY 324
>AW720070
Length = 484
Score = 25.4 bits (54), Expect = 3.6
Identities = 12/33 (36%), Positives = 19/33 (57%)
Frame = +2
Query: 105 SRKQLKERKNRAKKIRGVKKTKAADAAKAGKKK 137
+ K+ K++K + KK + VKK + K KKK
Sbjct: 104 AEKKRKKKKKKRKKKKQVKKESSQQQEKKKKKK 202
>TC12832 weakly similar to UP|POLX_TOBAC (P10978) Retrovirus-related Pol
polyprotein from transposon TNT 1-94 [Contains: Protease
; Reverse transcriptase ; Endonuclease] , partial (9%)
Length = 747
Score = 25.0 bits (53), Expect = 4.7
Identities = 14/43 (32%), Positives = 23/43 (52%)
Frame = +2
Query: 11 RKFMTNRLLSRKQFIVDVLHPGRANVSKAELKEKLARLYDVKD 53
++F N + ++ D+L G ELK +LAR +D+KD
Sbjct: 65 KRFDDNDFIILLLYVDDMLVVGPNKDRVQELKAQLAREFDMKD 193
>TC15643 similar to UP|O64720 (O64720) At2g02560 protein, partial (17%)
Length = 966
Score = 24.6 bits (52), Expect = 6.2
Identities = 16/70 (22%), Positives = 28/70 (39%)
Frame = +3
Query: 41 LKEKLARLYDVKDPNTVFVFKFRTHFGGGKSTGFGLIYDSLENAKKYEPKYRLIRNGLDT 100
LK L YDVK P + + K + + D L+ ++PK ++ +D
Sbjct: 342 LKSGLDDHYDVKMPCHLILSKLADNCPSAVLAVLDSLVDPLQKTINFKPKQDAVKQEVDR 521
Query: 101 KVEKSRKQLK 110
+ R L+
Sbjct: 522 NEDMIRSALR 551
>TC8122 similar to UP|Q94A43 (Q94A43) AT4g36780/C7A10_580, partial (74%)
Length = 1505
Score = 24.3 bits (51), Expect = 8.1
Identities = 14/38 (36%), Positives = 20/38 (51%)
Frame = +2
Query: 57 VFVFKFRTHFGGGKSTGFGLIYDSLENAKKYEPKYRLI 94
+++F T GGG STG + EN K+ E + R I
Sbjct: 140 LYIFFAGTMAGGGGSTGRLPTWKERENNKRRERRRRAI 253
>TC20230 homologue to UP|CYC_BRAOL (P00050) Cytochrome c, complete
Length = 598
Score = 24.3 bits (51), Expect = 8.1
Identities = 11/29 (37%), Positives = 17/29 (57%)
Frame = +3
Query: 77 IYDSLENAKKYEPKYRLIRNGLDTKVEKS 105
+YD L N KKY P +++ GL E++
Sbjct: 267 LYDYLLNPKKYIPGTKMVFPGLKKPQERA 353
>TC19312 similar to UP|Q9XIS4 (Q9XIS4) Branching enzyme 3 , partial (4%)
Length = 523
Score = 24.3 bits (51), Expect = 8.1
Identities = 10/25 (40%), Positives = 17/25 (68%)
Frame = -2
Query: 34 ANVSKAELKEKLARLYDVKDPNTVF 58
ANVS +LK +R ++ +DP+ +F
Sbjct: 213 ANVSMVQLKLLTSRSFEAEDPSGMF 139
>BP064293
Length = 505
Score = 24.3 bits (51), Expect = 8.1
Identities = 11/29 (37%), Positives = 17/29 (57%)
Frame = -1
Query: 77 IYDSLENAKKYEPKYRLIRNGLDTKVEKS 105
+YD L N KKY P +++ GL E++
Sbjct: 316 LYDYLLNPKKYIPGTKMVFPGLKKPQERA 230
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.318 0.134 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,492,015
Number of Sequences: 28460
Number of extensions: 14672
Number of successful extensions: 89
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 88
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 89
length of query: 137
length of database: 4,897,600
effective HSP length: 81
effective length of query: 56
effective length of database: 2,592,340
effective search space: 145171040
effective search space used: 145171040
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 50 (23.9 bits)
Lotus: description of TM0096b.1


