

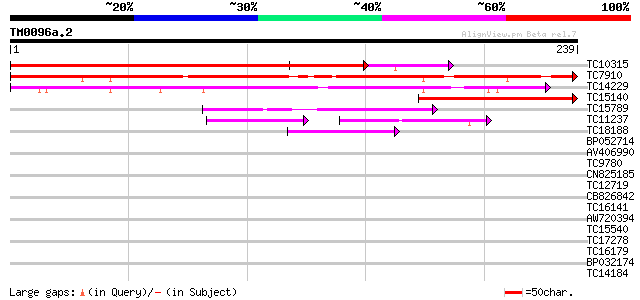
BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0096a.2
(239 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TC10315 similar to UP|P93166 (P93166) SCOF-1, partial (42%) 313 2e-86
TC7910 homologue to UP|P93166 (P93166) SCOF-1, partial (52%) 276 2e-75
TC14229 similar to UP|P93166 (P93166) SCOF-1, partial (38%) 202 5e-53
TC15140 similar to UP|P93166 (P93166) SCOF-1, partial (25%) 140 1e-34
TC15789 similar to UP|O22086 (O22086) ZPT2-14, partial (37%) 65 1e-11
TC11237 similar to UP|Q43614 (Q43614) DNA-binding protein, parti... 54 3e-08
TC18188 similar to UP|ZFP6_ARATH (Q39265) Zinc finger protein 6,... 39 6e-04
BP052714 35 0.009
AV406990 35 0.015
TC9780 similar to AAQ56830 (AAQ56830) At5g09670, partial (55%) 33 0.034
CN825185 33 0.034
TC12719 homologue to UP|ZFP1_ARATH (Q42485) Zinc finger protein ... 32 0.098
CB826842 32 0.098
TC16141 weakly similar to GB|AAC77862.2|20197419|AC005623 Argona... 31 0.17
AW720394 31 0.17
TC15540 similar to UP|Q9NGS5 (Q9NGS5) Prespore protein MF12 (Fra... 31 0.17
TC17278 similar to UP|Q8GT74 (Q8GT74) NEP1-interacting protein 2... 31 0.22
TC16179 weakly similar to UP|Q7PXR0 (Q7PXR0) AgCP12009, partial ... 30 0.37
BP032174 30 0.49
TC14184 similar to UP|Q93WZ6 (Q93WZ6) Abscisic stress ripening-l... 29 0.64
>TC10315 similar to UP|P93166 (P93166) SCOF-1, partial (42%)
Length = 592
Score = 313 bits (802), Expect = 2e-86
Identities = 151/151 (100%), Positives = 151/151 (100%)
Frame = +2
Query: 1 MAMEALNSPTTTAPSFTFNDHPTLRHPAEPWAKRKRSKRSRSCSEEEYLALCLIMLARGG 60
MAMEALNSPTTTAPSFTFNDHPTLRHPAEPWAKRKRSKRSRSCSEEEYLALCLIMLARGG
Sbjct: 140 MAMEALNSPTTTAPSFTFNDHPTLRHPAEPWAKRKRSKRSRSCSEEEYLALCLIMLARGG 319
Query: 61 AATTTATPPLQPATAPSGSSRLSYKCSVCDKAFPSYQALGGHKASHRKHSGGGGEDHSAG 120
AATTTATPPLQPATAPSGSSRLSYKCSVCDKAFPSYQALGGHKASHRKHSGGGGEDHSAG
Sbjct: 320 AATTTATPPLQPATAPSGSSRLSYKCSVCDKAFPSYQALGGHKASHRKHSGGGGEDHSAG 499
Query: 121 ATSSAVATTTSSANTGSGGKAHECSICHKSF 151
ATSSAVATTTSSANTGSGGKAHECSICHKSF
Sbjct: 500 ATSSAVATTTSSANTGSGGKAHECSICHKSF 592
Score = 52.8 bits (125), Expect = 5e-08
Identities = 29/72 (40%), Positives = 42/72 (58%), Gaps = 3/72 (4%)
Frame = +2
Query: 119 AGATSSAVATTTSSANTGSGGKAHECSICHKSFPTGQALGGHK---RCHYEGGGGSSSAT 175
A T++ ++A +GS +++CS+C K+FP+ QALGGHK R H GGG SA
Sbjct: 320 AATTTATPPLQPATAPSGSSRLSYKCSVCDKAFPSYQALGGHKASHRKHSGGGGEDHSAG 499
Query: 176 ASEGVGSTHTVS 187
A+ +T T S
Sbjct: 500 ATSSAVATTTSS 535
>TC7910 homologue to UP|P93166 (P93166) SCOF-1, partial (52%)
Length = 1035
Score = 276 bits (706), Expect = 2e-75
Identities = 158/252 (62%), Positives = 180/252 (70%), Gaps = 13/252 (5%)
Frame = +3
Query: 1 MAMEALNSPTTTAPSFTFNDHPTLRHPAE-PWAKRKRSKRSR--------SCSEEEYLAL 51
MAMEAL SPTT AP+FT P + + PWAKRKRSKRSR SC+EEEYLAL
Sbjct: 93 MAMEALKSPTTAAPTFTPFQEPNHSYMVDAPWAKRKRSKRSRTDSHHNHASCTEEEYLAL 272
Query: 52 CLIMLARGGAATTTATPPLQPATAPSGSSRLSYKCSVCDKAFPSYQALGGHKASHRKHSG 111
CLIMLARG A T +PA P + +LSYKCSVC+K FPSYQALGGHKASHRK +G
Sbjct: 273 CLIMLARGSTAVTPKLTLSRPA--PVTAEKLSYKCSVCEKTFPSYQALGGHKASHRKLAG 446
Query: 112 GGGEDHSAGATSSAVATTTSSANTGSGGKAHECSICHKSFPTGQALGGHKRCHYEGGGGS 171
EDHS TSSAV TTSSA+ G GGK H+CSIC KSFPTGQALGGHKRCHYEGGGG+
Sbjct: 447 AAAEDHS---TSSAV--TTSSASNG-GGKVHQCSICQKSFPTGQALGGHKRCHYEGGGGA 608
Query: 172 SS---ATASEGVGSTHTVSHSHHRDFDLNIPALPEFPSTK-AGEDEVESPHPVMKKKQRV 227
SS ATASEGVGST HSH R+FDLN+PA P+F ++K E+EV SP P K +
Sbjct: 609 SSTATATASEGVGST----HSHQRNFDLNLPAFPDFSASKFFVEEEVSSPLPSKKPR--- 767
Query: 228 FMIPKIEIPQLH 239
++PKIEIP +
Sbjct: 768 -LLPKIEIPHYY 800
>TC14229 similar to UP|P93166 (P93166) SCOF-1, partial (38%)
Length = 1175
Score = 202 bits (513), Expect = 5e-53
Identities = 129/280 (46%), Positives = 157/280 (56%), Gaps = 52/280 (18%)
Frame = +1
Query: 1 MAMEALNSPTT-TAP---SFTFNDHPTLRHPAEPWAKRKRSKRSR----SCSEEEYLALC 52
MA+E LNSPT T P S+ + H EPW K+KRSKR R + +EEEYLALC
Sbjct: 55 MALETLNSPTAATTPFRSSYQQEEEEEELHHHEPWTKKKRSKRPRFDNPTTTEEEYLALC 234
Query: 53 LIMLARGGAA------TTTATPPLQPATAPSGSS----------RLSYKCSVCDKAFPSY 96
LIMLA+ G T T L+ ++ + S +L++KCSVCDKAFPSY
Sbjct: 235 LIMLAQSGNNNNKNNNTDNNTNTLRAESSSNSQSQSPPQQQPPLKLTHKCSVCDKAFPSY 414
Query: 97 QALGGHKASHRKHSGGGGEDHSAGATSSAVATTTSSANTGSGGKAHECSICHKSFPTGQA 156
QALGGHKASHRK S +A +A +T+ TG+GGK HECSICHKSFPTGQA
Sbjct: 415 QALGGHKASHRKSSSENQTPAAAAVNDNASVSTS----TGNGGKMHECSICHKSFPTGQA 582
Query: 157 LGGHKRCHYEGGGGSSS--------------ATASEGVGSTHTVSHSHHRDFDLNIPA-L 201
LGGHKRCHYEGG S+S T+ G S H++ R FDLN+PA L
Sbjct: 583 LGGHKRCHYEGGKSSNSTNSNVHANTSSAVTTTSDGGAASAHSL-----RGFDLNLPAPL 747
Query: 202 PEF-------------PSTKAGEDEVESPHPVMKKKQRVF 228
EF ++GE EVESP PV K+ R+F
Sbjct: 748 TEFWTPLGIGGGDSRTNKIRSGEQEVESPLPVTAKRPRLF 867
>TC15140 similar to UP|P93166 (P93166) SCOF-1, partial (25%)
Length = 503
Score = 140 bits (354), Expect = 1e-34
Identities = 67/67 (100%), Positives = 67/67 (100%)
Frame = +1
Query: 173 SATASEGVGSTHTVSHSHHRDFDLNIPALPEFPSTKAGEDEVESPHPVMKKKQRVFMIPK 232
SATASEGVGSTHTVSHSHHRDFDLNIPALPEFPSTKAGEDEVESPHPVMKKKQRVFMIPK
Sbjct: 1 SATASEGVGSTHTVSHSHHRDFDLNIPALPEFPSTKAGEDEVESPHPVMKKKQRVFMIPK 180
Query: 233 IEIPQLH 239
IEIPQLH
Sbjct: 181 IEIPQLH 201
>TC15789 similar to UP|O22086 (O22086) ZPT2-14, partial (37%)
Length = 747
Score = 65.1 bits (157), Expect = 1e-11
Identities = 36/99 (36%), Positives = 52/99 (52%)
Frame = +1
Query: 82 LSYKCSVCDKAFPSYQALGGHKASHRKHSGGGGEDHSAGATSSAVATTTSSANTGSGGKA 141
+ ++C C++ F S+QALGGH+ASH H E+ + T S + +
Sbjct: 235 VEFECKTCNRKFSSFQALGGHRASHN-HKRVKLEEQAK----------TPSLWDNNKPRM 381
Query: 142 HECSICHKSFPTGQALGGHKRCHYEGGGGSSSATASEGV 180
H CS+C F GQALGGH R H G SSS+++S +
Sbjct: 382 HVCSVCGLGFSLGQALGGHMRKHRNNEGFSSSSSSSYSI 498
Score = 35.0 bits (79), Expect = 0.012
Identities = 17/42 (40%), Positives = 20/42 (47%)
Frame = +1
Query: 86 CSVCDKAFPSYQALGGHKASHRKHSGGGGEDHSAGATSSAVA 127
CSVC F QALGGH HR + G S+ + VA
Sbjct: 388 CSVCGLGFSLGQALGGHMRKHRNNEGFSSSSSSSYSIKEEVA 513
>TC11237 similar to UP|Q43614 (Q43614) DNA-binding protein, partial (20%)
Length = 523
Score = 53.5 bits (127), Expect = 3e-08
Identities = 29/66 (43%), Positives = 36/66 (53%), Gaps = 2/66 (3%)
Frame = +3
Query: 140 KAHECSICHKSFPTGQALGGHKRCHYEGGGGSSSATASEGVGSTHTVSHSHHR--DFDLN 197
K HECSIC F +GQALGGH R H +++ T S + ST V+ DLN
Sbjct: 48 KIHECSICGSEFTSGQALGGHMRRH-RASANAAAETNSNAITSTTVVARPPRNILQLDLN 224
Query: 198 IPALPE 203
+PA PE
Sbjct: 225 LPAPPE 242
Score = 40.8 bits (94), Expect = 2e-04
Identities = 18/43 (41%), Positives = 24/43 (54%)
Frame = +3
Query: 84 YKCSVCDKAFPSYQALGGHKASHRKHSGGGGEDHSAGATSSAV 126
++CS+C F S QALGGH HR + E +S TS+ V
Sbjct: 54 HECSICGSEFTSGQALGGHMRRHRASANAAAETNSNAITSTTV 182
>TC18188 similar to UP|ZFP6_ARATH (Q39265) Zinc finger protein 6, partial
(21%)
Length = 594
Score = 39.3 bits (90), Expect = 6e-04
Identities = 17/47 (36%), Positives = 25/47 (53%)
Frame = +3
Query: 118 SAGATSSAVATTTSSANTGSGGKAHECSICHKSFPTGQALGGHKRCH 164
S+ + + A + S NT SG + +C C + F QALGGH+ H
Sbjct: 237 SSDSGNGDAAGSASGINTPSGDRKKKCQYCCREFANSQALGGHQNAH 377
Score = 33.9 bits (76), Expect = 0.026
Identities = 24/69 (34%), Positives = 30/69 (42%), Gaps = 21/69 (30%)
Frame = +3
Query: 61 AATTTATPPLQP--ATAPSGSSRLSY-------------------KCSVCDKAFPSYQAL 99
AA TP P AT+PSGSS KC C + F + QAL
Sbjct: 177 AAALEDTPDSLPVKATSPSGSSDSGNGDAAGSASGINTPSGDRKKKCQYCCREFANSQAL 356
Query: 100 GGHKASHRK 108
GGH+ +H+K
Sbjct: 357 GGHQNAHKK 383
>BP052714
Length = 511
Score = 35.4 bits (80), Expect = 0.009
Identities = 18/50 (36%), Positives = 26/50 (52%), Gaps = 1/50 (2%)
Frame = -1
Query: 58 RGGAATTTATPPLQPATAPSGSSRLSYKCSVCDKAFPSYQALGGH-KASH 106
+ G A+ + P +P T SG Y C C+++F + ALG H KA H
Sbjct: 325 QAGKASASTKQPAKPQTPKSGGE---YSCKPCNRSFKTEDALGSHXKAKH 185
>AV406990
Length = 392
Score = 34.7 bits (78), Expect = 0.015
Identities = 23/75 (30%), Positives = 36/75 (47%)
Frame = +1
Query: 145 SICHKSFPTGQALGGHKRCHYEGGGGSSSATASEGVGSTHTVSHSHHRDFDLNIPALPEF 204
++ + FPTG+ ++ EG G+S TA E + +SHSHH +L +P P
Sbjct: 70 TLLRRHFPTGRVPNPDQKT--EGEAGASPKTAPE---AHPPISHSHHSTTNLTLPPPPLP 234
Query: 205 PSTKAGEDEVESPHP 219
T + + SP P
Sbjct: 235 SQTPWMQISMPSPSP 279
>TC9780 similar to AAQ56830 (AAQ56830) At5g09670, partial (55%)
Length = 1250
Score = 33.5 bits (75), Expect = 0.034
Identities = 29/88 (32%), Positives = 36/88 (39%), Gaps = 11/88 (12%)
Frame = +2
Query: 111 GGGGEDHSAGATSSAVATTTSSANTGSGGKAHECSICHK----SFPTGQALGGHKRCHYE 166
GGG G + A +T G GGK + C K S P +A GG KRC Y
Sbjct: 818 GGGRRCQYLGCSKGAQGSTMFCKAHG-GGKRCSFAGCSKGAEGSTPLCKAHGGGKRCLYN 994
Query: 167 GGG-------GSSSATASEGVGSTHTVS 187
GGG G ++ + G G VS
Sbjct: 995 GGGICGKSVHGGTNFCVAHGGGKRCAVS 1078
>CN825185
Length = 541
Score = 33.5 bits (75), Expect = 0.034
Identities = 13/24 (54%), Positives = 16/24 (66%)
Frame = +3
Query: 84 YKCSVCDKAFPSYQALGGHKASHR 107
Y C C K F S QA+GGH+ +HR
Sbjct: 198 YGCLYCSKRFSSVQAVGGHQNAHR 269
Score = 30.0 bits (66), Expect = 0.37
Identities = 18/58 (31%), Positives = 26/58 (44%), Gaps = 12/58 (20%)
Frame = +3
Query: 119 AGATSSAVATTTSSANTGS------------GGKAHECSICHKSFPTGQALGGHKRCH 164
A A+SS V++ +S T G + C C K F + QA+GGH+ H
Sbjct: 93 AKASSSPVSSGENSITTRKRQMGMWLSLSQPGSHGYGCLYCSKRFSSVQAVGGHQNAH 266
>TC12719 homologue to UP|ZFP1_ARATH (Q42485) Zinc finger protein 1, partial
(18%)
Length = 548
Score = 32.0 bits (71), Expect = 0.098
Identities = 13/38 (34%), Positives = 20/38 (52%)
Frame = +2
Query: 140 KAHECSICHKSFPTGQALGGHKRCHYEGGGGSSSATAS 177
+ C+ CH+ F + QAL G +C EG S ++S
Sbjct: 395 RVFSCNYCHRKFYSSQALWGSPKCTQEGEVNSKKKSSS 508
>CB826842
Length = 637
Score = 32.0 bits (71), Expect = 0.098
Identities = 13/59 (22%), Positives = 26/59 (44%)
Frame = +1
Query: 140 KAHECSICHKSFPTGQALGGHKRCHYEGGGGSSSATASEGVGSTHTVSHSHHRDFDLNI 198
+++ C+ C + F QALGGH H ++ + ++ + H D + N+
Sbjct: 121 RSYSCTFCKRGFSNAQALGGHMNIHTRDRAKLKQSSEENLLSLDISMKSTEHLDSEENL 297
Score = 32.0 bits (71), Expect = 0.098
Identities = 12/26 (46%), Positives = 16/26 (61%)
Frame = +1
Query: 83 SYKCSVCDKAFPSYQALGGHKASHRK 108
SY C+ C + F + QALGGH H +
Sbjct: 124 SYSCTFCKRGFSNAQALGGHMNIHTR 201
>TC16141 weakly similar to GB|AAC77862.2|20197419|AC005623 Argonaute
(AGO1)-like protein {Arabidopsis thaliana;} , partial
(14%)
Length = 598
Score = 31.2 bits (69), Expect = 0.17
Identities = 17/30 (56%), Positives = 19/30 (62%)
Frame = -1
Query: 110 SGGGGEDHSAGATSSAVATTTSSANTGSGG 139
SGGG +AG+TSSA TTS TG GG
Sbjct: 184 SGGG----AAGSTSSAFRGTTSEGRTGGGG 107
>AW720394
Length = 587
Score = 31.2 bits (69), Expect = 0.17
Identities = 20/54 (37%), Positives = 29/54 (53%), Gaps = 5/54 (9%)
Frame = +2
Query: 8 SPTTTAPSFTFNDHPTLRHPAEPWAKRK-RSKRSRSCSE----EEYLALCLIML 56
SPTT+ PS T +D P W++RK S+R + CS E + L L++L
Sbjct: 233 SPTTSTPSTTTSDPWRCEVPRNLWSRRKPYSRRIQDCSTLKMMREMMLLMLMLL 394
>TC15540 similar to UP|Q9NGS5 (Q9NGS5) Prespore protein MF12 (Fragment),
partial (6%)
Length = 806
Score = 31.2 bits (69), Expect = 0.17
Identities = 27/99 (27%), Positives = 41/99 (41%)
Frame = +1
Query: 61 AATTTATPPLQPATAPSGSSRLSYKCSVCDKAFPSYQALGGHKASHRKHSGGGGEDHSAG 120
AATTTAT PA + ++ + S AFP A ++ + +A
Sbjct: 28 AATTTATTTTYPAPTTTAAATST---SPTAAAFPDAAATTPSTSATTTLAAAAAA--TAS 192
Query: 121 ATSSAVATTTSSANTGSGGKAHECSICHKSFPTGQALGG 159
A ++ A+TTSS G + SIC +S+ G
Sbjct: 193 AVAAITASTTSSTAATDGWGRNGSSICSRSWAVTNGFSG 309
>TC17278 similar to UP|Q8GT74 (Q8GT74) NEP1-interacting protein 2, partial
(62%)
Length = 610
Score = 30.8 bits (68), Expect = 0.22
Identities = 13/48 (27%), Positives = 25/48 (52%), Gaps = 1/48 (2%)
Frame = +3
Query: 119 AGATSSAVATTTSSANTG-SGGKAHECSICHKSFPTGQALGGHKRCHY 165
+GA+ + + T + N G + G+ CS+C + F G+ + CH+
Sbjct: 240 SGASVAKIPQVTITGNNGDASGQRDSCSVCLQDFQLGETVRSLPYCHH 383
>TC16179 weakly similar to UP|Q7PXR0 (Q7PXR0) AgCP12009, partial (20%)
Length = 814
Score = 30.0 bits (66), Expect = 0.37
Identities = 16/36 (44%), Positives = 18/36 (49%)
Frame = -1
Query: 7 NSPTTTAPSFTFNDHPTLRHPAEPWAKRKRSKRSRS 42
+S T AP NDHP L EPW +R R RS
Sbjct: 337 DSTRTNAP----NDHPNLSEHCEPW*QRS*RSRKRS 242
>BP032174
Length = 522
Score = 29.6 bits (65), Expect = 0.49
Identities = 26/88 (29%), Positives = 32/88 (35%), Gaps = 1/88 (1%)
Frame = -2
Query: 8 SPTTTAPSFTFNDHPTLRHPAEPWAKRKRSKRSRSCSEEEYLALCLIMLARGGAATTTAT 67
SPTT S D P P P A K + S L L M R + T
Sbjct: 521 SPTTAPTSTASQDPPASGTPKSPLASAKS*GSASGKSASPVFTLTLAMSFRAPSTTA*WF 342
Query: 68 PPLQ-PATAPSGSSRLSYKCSVCDKAFP 94
PP P+ AP+ S + S+C P
Sbjct: 341 PPCSIPSNAPASRSP-ALTVSLCASHLP 261
>TC14184 similar to UP|Q93WZ6 (Q93WZ6) Abscisic stress ripening-like
protein, partial (57%)
Length = 1141
Score = 29.3 bits (64), Expect = 0.64
Identities = 31/111 (27%), Positives = 48/111 (42%)
Frame = +3
Query: 71 QPATAPSGSSRLSYKCSVCDKAFPSYQALGGHKASHRKHSGGGGEDHSAGATSSAVATTT 130
+P +G ++ SY S + + Y + G+K S + SGGG E TSS +T
Sbjct: 135 RPTDTETGYNKTSY--STDEPSGGDYDS--GYKKSSYETSGGGYET-GYNKTSSY---ST 290
Query: 131 SSANTGSGGKAHECSICHKSFPTGQALGGHKRCHYEGGGGSSSATASEGVG 181
N+G GG ++ ++ T + GG GGG S+ G G
Sbjct: 291 DEPNSGGGG--YDSGYNKTAYSTDEPSGGGYGTGGAGGGYSTGGGYGTGGG 437
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.312 0.126 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,858,368
Number of Sequences: 28460
Number of extensions: 77433
Number of successful extensions: 641
Number of sequences better than 10.0: 85
Number of HSP's better than 10.0 without gapping: 607
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 620
length of query: 239
length of database: 4,897,600
effective HSP length: 87
effective length of query: 152
effective length of database: 2,421,580
effective search space: 368080160
effective search space used: 368080160
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 54 (25.4 bits)
Lotus: description of TM0096a.2


