

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0094b.11
(1170 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
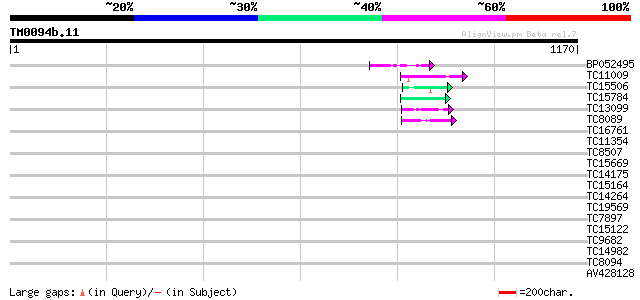
Score E
Sequences producing significant alignments: (bits) Value
BP052495 59 4e-09
TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Ar... 45 7e-05
TC15506 similar to UP|Q9SEE9 (Q9SEE9) Arginine/serine-rich prote... 45 7e-05
TC15784 similar to UP|PE31_ARATH (Q9LHA7) Peroxidase 31 precurso... 44 1e-04
TC13099 weakly similar to UP|CBL9_ARATH (Q9FJ13) COBRA-like prot... 44 2e-04
TC8089 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabinog... 43 3e-04
TC16761 similar to GB|AAL32703.1|17065098|AY062625 nucleotide su... 40 0.002
TC11354 similar to UP|CAE54480 (CAE54480) Alpha glucosidase II ... 40 0.002
TC8507 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetyla... 40 0.003
TC15669 homologue to UP|PRS6_ARATH (Q9SEI4) 26S protease regulat... 40 0.003
TC14175 similar to UP|Q9SII5 (Q9SII5) At2g17230 protein (At2g172... 40 0.003
TC15164 similar to UP|Q9ZTK7 (Q9ZTK7) CONSTANS-like protein 2, p... 39 0.005
TC14264 similar to UP|Q84NF9 (Q84NF9) Histone H1 (Fragment), par... 39 0.005
TC19569 similar to UP|AAR11301 (AAR11301) Lectin-like receptor k... 39 0.006
TC7897 weakly similar to UP|ENL2_ARATH (Q9T076) Early nodulin-li... 39 0.006
TC15122 similar to UP|Q94AY5 (Q94AY5) At1g19950/T20H2_25, partia... 38 0.008
TC9682 similar to UP|Q8VZC0 (Q8VZC0) DTDP-glucose 4-6-dehydratas... 38 0.011
TC14982 similar to UP|Q9FPI2 (Q9FPI2) At1g17620, partial (32%) 37 0.014
TC8094 homologue to UP|EFTU_PEA (O24310) Elongation factor Tu, c... 37 0.018
AV428128 36 0.031
>BP052495
Length = 524
Score = 58.9 bits (141), Expect = 4e-09
Identities = 54/138 (39%), Positives = 68/138 (49%), Gaps = 3/138 (2%)
Frame = -3
Query: 742 VSLADKL---KSRKRKPKPVASASQKRAKGAGVSTTSGASKLASDAQPEVDDKEGGGDAA 798
V L D L +SRKR P SASQKRAKGAG + S S +Q E + + GG+
Sbjct: 429 VYLCDHLLLFQSRKRGATPTTSASQKRAKGAGEPSAGNPSPAKSASQRE--EVQQGGEQG 256
Query: 799 IQADVHEHSTSNPPSRENSPAPNPAKSKAEGVEKPPKAASSAKKTSTSEGRRKVRSKSGH 858
+ + SNP + +P P VE + AKKTS G+R R KSG
Sbjct: 255 V-----DEQASNPLLQGTTPIP---------VE-----MAQAKKTS---GKRYSR-KSGS 145
Query: 859 KSPRRSRSSTNSSPSKES 876
S S+SST+SSP K S
Sbjct: 144 SSSHHSKSSTSSSPLKAS 91
Score = 28.9 bits (63), Expect = 5.0
Identities = 12/18 (66%), Positives = 15/18 (82%)
Frame = -1
Query: 735 DDDEDDDVSLADKLKSRK 752
+DD+DDDV+L DKLK K
Sbjct: 524 EDDDDDDVALPDKLKIPK 471
>TC11009 similar to GB|AAP21377.1|30102918|BT006569 At1g47970 {Arabidopsis
thaliana;}, partial (44%)
Length = 700
Score = 45.1 bits (105), Expect = 7e-05
Identities = 40/150 (26%), Positives = 61/150 (40%), Gaps = 11/150 (7%)
Frame = -2
Query: 807 STSNPPSRENS-PAPN----------PAKSKAEGVEKPPKAASSAKKTSTSEGRRKVRSK 855
S+S+PPS E+S P P+ PA S + P ++S +K S +
Sbjct: 435 SSSSPPSSESSSPEPSLSDLFLFDGTPAFSPSSLSLSSPSSSSGSKSLDASSSSAAATVR 256
Query: 856 SGHKSPRRSRSSTNSSPSKESACQETQGAISPIREAEPLPGKDDNPMPPPKSTATVQPSP 915
S +S SS++SS S S+ + G+ P R P + S+++ P P
Sbjct: 255 RTRYSVPKSSSSSSSSSSSSSSSSSSSGSPPPPRPPSTPPSSSSS-----SSSSSSSPPP 91
Query: 916 QAKGGSSSHVSSEKLDTLFEEDPLAALDGF 945
A SSS SS + PL + F
Sbjct: 90 GASSSSSSSPSSSSPSSSSSSSPLILVREF 1
Score = 33.1 bits (74), Expect = 0.26
Identities = 21/82 (25%), Positives = 35/82 (42%), Gaps = 8/82 (9%)
Frame = +2
Query: 723 GQGQASRDEDDDDDDEDDDVSLADKLKSRKRKPKPVASASQKRAKG--------AGVSTT 774
G G D+DDDDDD+D++ + L + + VA+A + A +
Sbjct: 158 GGGGDPDDDDDDDDDDDEEEEEEEDLGTEYLVRRTVAAAEDEEASSDFEPEEDEGDDNDN 337
Query: 775 SGASKLASDAQPEVDDKEGGGD 796
K ++ + DK+G GD
Sbjct: 338 DDGEKAGVPSKRKRSDKDGSGD 403
Score = 31.6 bits (70), Expect = 0.76
Identities = 22/98 (22%), Positives = 39/98 (39%)
Frame = -2
Query: 759 ASASQKRAKGAGVSTTSGASKLASDAQPEVDDKEGGGDAAIQADVHEHSTSNPPSRENSP 818
+S+S ++ A S+ + + + P+ ++ + S S PP R S
Sbjct: 318 SSSSGSKSLDASSSSAAATVRRTRYSVPKSSSSSSSSSSSSSSS-SSSSGSPPPPRPPST 142
Query: 819 APNPAKSKAEGVEKPPKAASSAKKTSTSEGRRKVRSKS 856
P+ + S + PP ASS+ +S S S S
Sbjct: 141 PPSSSSSSSSSSSSPPPGASSSSSSSPSSSSPSSSSSS 28
Score = 29.6 bits (65), Expect = 2.9
Identities = 25/106 (23%), Positives = 45/106 (41%)
Frame = -2
Query: 772 STTSGASKLASDAQPEVDDKEGGGDAAIQADVHEHSTSNPPSRENSPAPNPAKSKAEGVE 831
S+ S +S +S +D A ++ + S+ S +S + + + S + G
Sbjct: 342 SSLSLSSPSSSSGSKSLDASSSSAAATVRRTRYSVPKSSSSSSSSSSSSS-SSSSSSGSP 166
Query: 832 KPPKAASSAKKTSTSEGRRKVRSKSGHKSPRRSRSSTNSSPSKESA 877
PP+ S+ +S+S G S S S ++SSPS S+
Sbjct: 165 PPPRPPSTPPSSSSSSSSSSSSPPPGASSSSSS-SPSSSSPSSSSS 31
Score = 29.6 bits (65), Expect = 2.9
Identities = 12/14 (85%), Positives = 12/14 (85%)
Frame = +2
Query: 728 SRDEDDDDDDEDDD 741
S DEDDDDD EDDD
Sbjct: 20 SGDEDDDDDGEDDD 61
>TC15506 similar to UP|Q9SEE9 (Q9SEE9) Arginine/serine-rich protein, partial
(24%)
Length = 677
Score = 45.1 bits (105), Expect = 7e-05
Identities = 33/113 (29%), Positives = 44/113 (38%), Gaps = 10/113 (8%)
Frame = +2
Query: 811 PPSRENSPAPNPAKSKAEGVEKPPKAASSAKKTSTSEGRRKVRSKSGHKSPRRSRS---- 866
PP R SP P A+S PP+ + ++ S S RR RS S SPRR R
Sbjct: 53 PPPRRRSPPPRRARS-------PPRRSPIGRRRSRSPIRRPARSNSRSFSPRRGRPPVRR 211
Query: 867 ------STNSSPSKESACQETQGAISPIREAEPLPGKDDNPMPPPKSTATVQP 913
S + SP K S ++ P+R + PPP +P
Sbjct: 212 GRSSSFSDSPSPRKVSRRSRSRSPRRPLRGGRASSNSSSSSSPPPPPPPARKP 370
>TC15784 similar to UP|PE31_ARATH (Q9LHA7) Peroxidase 31 precursor (Atperox
P31) (ATP41) , partial (38%)
Length = 475
Score = 44.3 bits (103), Expect = 1e-04
Identities = 29/108 (26%), Positives = 42/108 (38%), Gaps = 4/108 (3%)
Frame = +1
Query: 806 HSTSNPPSRENSP----APNPAKSKAEGVEKPPKAASSAKKTSTSEGRRKVRSKSGHKSP 861
HSTS NSP P+P PP++ASS+ ST S S
Sbjct: 112 HSTSTNTRAHNSPKSSATPSPTNKSPPPPPAPPRSASSSTTASTPTAATPPSSSPQLPST 291
Query: 862 RRSRSSTNSSPSKESACQETQGAISPIREAEPLPGKDDNPMPPPKSTA 909
+ + + T++SPS + + P P P PPP +T+
Sbjct: 292 KPNATPTSTSPSPATPSTSSSELKPPSNSLAPTPSPAPTSSPPPPATS 435
Score = 31.2 bits (69), Expect = 1.00
Identities = 32/122 (26%), Positives = 48/122 (39%)
Frame = +1
Query: 804 HEHSTSNPPSRENSPAPNPAKSKAEGVEKPPKAASSAKKTSTSEGRRKVRSKSGHKSPRR 863
H HS+S+ +SP + S + +A +S K ++T K S +P R
Sbjct: 46 HHHSSSSSQHSHSSPPSHTRDSHSTSTNT--RAHNSPKSSATPSPTNK--SPPPPPAPPR 213
Query: 864 SRSSTNSSPSKESACQETQGAISPIREAEPLPGKDDNPMPPPKSTATVQPSPQAKGGSSS 923
S SS+ ++ T A +P + LP N P S PSP SSS
Sbjct: 214 SASSSTTA--------STPTAATPPSSSPQLPSTKPNATPTSTS-----PSPATPSTSSS 354
Query: 924 HV 925
+
Sbjct: 355 EL 360
>TC13099 weakly similar to UP|CBL9_ARATH (Q9FJ13) COBRA-like protein 9
precursor, partial (14%)
Length = 426
Score = 43.5 bits (101), Expect = 2e-04
Identities = 35/107 (32%), Positives = 49/107 (45%)
Frame = +3
Query: 809 SNPPSRENSPAPNPAKSKAEGVEKPPKAASSAKKTSTSEGRRKVRSKSGHKSPRRSRSST 868
S+ R PNPA + PP ASS ++TS + R R+ S +SP + +ST
Sbjct: 48 SSSSQRRRLRRPNPATVYSSPTPTPP--ASSYRRTSPT--LRSSRTSSSPRSPCSTTAST 215
Query: 869 NSSPSKESACQETQGAISPIREAEPLPGKDDNPMPPPKSTATVQPSP 915
+SSP + S+ T + SP P +P P ATV SP
Sbjct: 216 SSSPGRSSSGFSTMSSWSP-----PPMRCSPTGLPSPPPLATVPSSP 341
>TC8089 similar to UP|FLA1_ARATH (Q9FM65) Fasciclin-like arabinogalactan
protein 1 precursor, partial (43%)
Length = 890
Score = 43.1 bits (100), Expect = 3e-04
Identities = 30/114 (26%), Positives = 47/114 (40%)
Frame = +2
Query: 809 SNPPSRENSPAPNPAKSKAEGVEKPPKAASSAKKTSTSEGRRKVRSKSGHKSPRRSRSST 868
S+PPS SP+P G + P +++ T++S+ H+SP S
Sbjct: 263 SSPPSTTTSPSPTSPPRSTNGPQSPSAPSTTPPWTTSSQNT--------HQSP----PSR 406
Query: 869 NSSPSKESACQETQGAISPIREAEPLPGKDDNPMPPPKSTATVQPSPQAKGGSS 922
SSPS S+ + + A P P + P PP++ SP + G S
Sbjct: 407 TSSPSTSSSTTSAPRSSTRSPTAPPSPPQCTKPPAPPRAPPDSSTSPTSAAGKS 568
Score = 30.8 bits (68), Expect = 1.3
Identities = 35/125 (28%), Positives = 47/125 (37%), Gaps = 16/125 (12%)
Frame = +2
Query: 807 STSNPPSRENSPAPNPAKSKAEGVEKPPKAA--SSAKKTSTSEGRRKVRSKSGHKSPRRS 864
+TS P S SP P+ + PP+A SS TS + + SP RS
Sbjct: 434 TTSAPRSSTRSPTAPPSPPQCTKPPAPPRAPPDSSTSPTSAAGKSDSAPRTTTAPSPLRS 613
Query: 865 ----RSSTNSSPSKESA-----CQETQGAISPIREAEP-----LPGKDDNPMPPPKSTAT 910
R S +SPS S Q+ P+ P + + PP+ T T
Sbjct: 614 SNP*RKSPTTSPSSRSVRFFPPPQQKPLHPHPLSRISPPLCPSTVARSSPTLSPPRRTRT 793
Query: 911 VQPSP 915
QPSP
Sbjct: 794 -QPSP 805
>TC16761 similar to GB|AAL32703.1|17065098|AY062625 nucleotide sugar
epimerase-like protein {Arabidopsis thaliana;}, partial
(40%)
Length = 794
Score = 40.4 bits (93), Expect = 0.002
Identities = 36/124 (29%), Positives = 49/124 (39%), Gaps = 3/124 (2%)
Frame = +1
Query: 806 HSTSNPPSRENSPAPNPAKSKAEGVEKPPKAASSAKKTSTSEGRRKVRSKSGHKSPRRSR 865
H T P N KS A+ P +SSA +S+ H S S
Sbjct: 181 HQTQAKP*SWNVTTATSVKSTAQSFSTLPPNSSSAPPSSS------------HSSSSSSS 324
Query: 866 SSTN--SSPSKESACQETQGAISPIREAEPLPGKDDNPMPPPKSTATVQPSPQ-AKGGSS 922
ST S P+ + T + P AEP PG+ + PPP + T PS A+ SS
Sbjct: 325 PSTTLPSPPTPPTTSTPTPTSSPPHSAAEP-PGRSKSATPPPHAAPTACPSSSPAQSASS 501
Query: 923 SHVS 926
+H +
Sbjct: 502 AHTA 513
>TC11354 similar to UP|CAE54480 (CAE54480) Alpha glucosidase II , partial
(6%)
Length = 583
Score = 40.4 bits (93), Expect = 0.002
Identities = 34/149 (22%), Positives = 56/149 (36%), Gaps = 6/149 (4%)
Frame = -1
Query: 781 ASDAQPEVDDKEGGGDAAIQADVHEHSTSNP-PSRENSPAPNPAKSKAEGVEKPPKAASS 839
+S A P ++ + AI+ HS S P P+ P +P S + PP ++ +
Sbjct: 445 SSSAPPSSPGRKKSSETAIRP----HSASEPDPAPPPPPPSSPPMSPSPTATSPPTSSPN 278
Query: 840 AKKTST-----SEGRRKVRSKSGHKSPRRSRSSTNSSPSKESACQETQGAISPIREAEPL 894
+ S + S ++ ST P+ ES + + P +
Sbjct: 277 THPPTPTPNP*SSPSPSTTTASSASPSTKTLPSTRPRPASESPTSSSPSSPPPSSGSSVS 98
Query: 895 PGKDDNPMPPPKSTATVQPSPQAKGGSSS 923
P + P PPP ++ T P A SSS
Sbjct: 97 PPRISPPPPPPSNSPTATPPSSATNPSSS 11
Score = 38.1 bits (87), Expect = 0.008
Identities = 34/118 (28%), Positives = 46/118 (38%)
Frame = -1
Query: 808 TSNPPSRENSPAPNPAKSKAEGVEKPPKAASSAKKTSTSEGRRKVRSKSGHKSPRRSRSS 867
TS+P + +P PNP S + A+ S K ++ R S S
Sbjct: 292 TSSPNTHPPTPTPNP*SSPSPSTTTASSASPSTKTLPSTRPRPA------------SESP 149
Query: 868 TNSSPSKESACQETQGAISPIREAEPLPGKDDNPMPPPKSTATVQPSPQAKGGSSSHV 925
T+SSPS S + ++SP R + P P PP S PS SSS V
Sbjct: 148 TSSSPS--SPPPSSGSSVSPPRISPP-------PPPPSNSPTATPPSSATNPSSSSSV 2
>TC8507 similar to UP|Q8LJS2 (Q8LJS2) Nucleolar histone deacetylase
HD2-P39, partial (40%)
Length = 620
Score = 39.7 bits (91), Expect = 0.003
Identities = 31/145 (21%), Positives = 55/145 (37%), Gaps = 7/145 (4%)
Frame = +3
Query: 728 SRDEDDDDDDEDDDVSLADKLKSR-------KRKPKPVASASQKRAKGAGVSTTSGASKL 780
S DEDD+DDD D+++ AD + P A +KRA + + T K
Sbjct: 18 SDDEDDEDDDSDEEMDDADSDSDESDSDDTDEETPVKKADQGKKRANDSALKTPVSTKKA 197
Query: 781 ASDAQPEVDDKEGGGDAAIQADVHEHSTSNPPSRENSPAPNPAKSKAEGVEKPPKAASSA 840
+ + D K+ G ++ P+P K + E + S
Sbjct: 198 KNITPEKTDGKKTG---------------------HTATPHPGKKGGKNSESKAQTPKSG 314
Query: 841 KKTSTSEGRRKVRSKSGHKSPRRSR 865
+ S S + S+SG + ++++
Sbjct: 315 GQFSCSPCSKSFNSESGFQQHKKAK 389
>TC15669 homologue to UP|PRS6_ARATH (Q9SEI4) 26S protease regulatory subunit
6B homolog (26S proteasome AAA-ATPase subunit RPT3)
(Regulatory particle triple-A ATPase subunit 3), partial
(34%)
Length = 608
Score = 39.7 bits (91), Expect = 0.003
Identities = 37/121 (30%), Positives = 52/121 (42%), Gaps = 12/121 (9%)
Frame = +3
Query: 806 HSTSNPPSRENSPAPNPAKSKAEGVEKPPKAASSA--KKTSTSEGRRKVRSKSGHKSPRR 863
H + +R SPAP P K+ + + A+SS+ K ST R++ S S RR
Sbjct: 141 HRPFSQRNRITSPAPTPMKTTSTAA*RRCSASSSSSISKRST*RRSRRI*SASFSARRRR 320
Query: 864 SRSSTN---SSPSKESACQETQGAISPIREAEPLPG-------KDDNPMPPPKSTATVQP 913
SR S SS S T +++P R+ G + +PP STAT P
Sbjct: 321 SRGSNRFPLSSASSWRWLTRTTASLAPQRDPTTTFGSSVLLTVSSSSLLPPSPSTATPTP 500
Query: 914 S 914
S
Sbjct: 501 S 503
>TC14175 similar to UP|Q9SII5 (Q9SII5) At2g17230 protein
(At2g17230/T23A1.9), partial (82%)
Length = 1447
Score = 39.7 bits (91), Expect = 0.003
Identities = 41/151 (27%), Positives = 57/151 (37%), Gaps = 32/151 (21%)
Frame = +1
Query: 804 HEHSTSNPPSRENSPA----------------PNPAKSKAEGVEKPPKAASSAKKTSTSE 847
H HSTS+ PS +SP P+ PP+ S A +TS S
Sbjct: 82 HRHSTSSSPSSSSSPPQLMERFKH*TSTTLQNSTPSSHSHPSEPSPPQNGSKAPQTSLSS 261
Query: 848 GR--------RKVRSKSG---HKSPRR--SRSSTNSSPSKESACQ--ETQGAISPIREAE 892
R + SG SPR SR+S++ SP ES + +T G +SP +
Sbjct: 262 STTWVLSSLPRSTSTSSGTATGPSPRSSLSRTSSDQSPPPESPPRR*QTGGVLSPSTPTK 441
Query: 893 P-LPGKDDNPMPPPKSTATVQPSPQAKGGSS 922
P L +P P T ++G S
Sbjct: 442 PALTSPAQSPSPASMPTGNTPTGDTSRGCQS 534
>TC15164 similar to UP|Q9ZTK7 (Q9ZTK7) CONSTANS-like protein 2, partial
(57%)
Length = 1185
Score = 38.9 bits (89), Expect = 0.005
Identities = 39/129 (30%), Positives = 54/129 (41%), Gaps = 13/129 (10%)
Frame = +2
Query: 807 STSNPPSRE--NSPAPNPAKSKAEGVEKPPKAASSAKKTSTSEGRRKVRSKSGHKSPRRS 864
S + PPS +SP A A KPP S A+ T + + P S
Sbjct: 164 SPATPPSTPPTSSPPATLASLSARSASKPPPP-SPARLTPLPSASPVTTTST---PPTPS 331
Query: 865 RSSTNSSPSKESACQETQGAISPI--------REAEPLPGKDDNPMPPPK---STATVQP 913
++TN+SPS+ S T SP + +PLPG P+P PK ST P
Sbjct: 332 PAATNASPSRRSTTTTTTPPTSPTPMLLTSPPMKLKPLPGY-FLPLPTPKAPISTPLTTP 508
Query: 914 SPQAKGGSS 922
+P++ SS
Sbjct: 509 TPKSTPSSS 535
>TC14264 similar to UP|Q84NF9 (Q84NF9) Histone H1 (Fragment), partial (76%)
Length = 1277
Score = 38.9 bits (89), Expect = 0.005
Identities = 38/157 (24%), Positives = 61/157 (38%), Gaps = 5/157 (3%)
Frame = +2
Query: 745 ADKLKSRKRKPKPVASASQKRAKGAGVSTTSGASKLASDAQPEVDDKEGGGDAAIQAD-- 802
A K K+ K PK A+A++ AK + +K A+ A+P K A +A
Sbjct: 563 ATKAKA-KPAPKSKAAATKTTAKAKPAAAAKPKAKPAAKAKPAAKAKAVAAPAKAKASPA 739
Query: 803 ---VHEHSTSNPPSRENSPAPNPAKSKAEGVEKPPKAASSAKKTSTSEGRRKVRSKSGHK 859
S + P N P P K+K KP + A +TST K
Sbjct: 740 KPKAKAKSKTAPRMNVNPPVP---KAKPAAKAKPAARPAKASRTSTRTTPGKKVPPPPKP 910
Query: 860 SPRRSRSSTNSSPSKESACQETQGAISPIREAEPLPG 896
+P+++ + +P+K + + SP + P G
Sbjct: 911 APKKAATPVKKAPAKSGKAKTVK---SPAKRTAPQRG 1012
Score = 34.7 bits (78), Expect = 0.090
Identities = 29/135 (21%), Positives = 55/135 (40%), Gaps = 14/135 (10%)
Frame = +2
Query: 807 STSNPPSRENSPAPNPAKS--KAEGVEKPPKAASSAK-------KTSTSEGRRKVRSKSG 857
+ S P + + P P AKS K + KP AA+ AK K + ++ K + +
Sbjct: 467 AASKPAAAKKKPQPAAAKSKPKPKAKAKPAAAATKAKAKPAPKSKAAATKTTAKAKPAAA 646
Query: 858 HKSPRRSRSSTNSSPSKESACQETQGAISPIR-----EAEPLPGKDDNPMPPPKSTATVQ 912
K + + + ++ + SP + +++ P + NP P PK+ +
Sbjct: 647 AKPKAKPAAKAKPAAKAKAVAAPAKAKASPAKPKAKAKSKTAPRMNVNP-PVPKAKPAAK 823
Query: 913 PSPQAKGGSSSHVSS 927
P A+ +S S+
Sbjct: 824 AKPAARPAKASRTST 868
>TC19569 similar to UP|AAR11301 (AAR11301) Lectin-like receptor kinase 1;1,
partial (16%)
Length = 486
Score = 38.5 bits (88), Expect = 0.006
Identities = 32/123 (26%), Positives = 49/123 (39%), Gaps = 14/123 (11%)
Frame = +1
Query: 807 STSNPPSRENSPAPNPA----------KSKAEGVEKPPKAASSAKKTSTSEGRRKVR--- 853
S S PP P +P+ S + E PP A S++ K+ TS +
Sbjct: 64 SYSPPPPPSQQPTHSPSTSPTSTTTPPPSPTKVTENPPTAPSTSTKSPTSSASEEPSPPN 243
Query: 854 -SKSGHKSPRRSRSSTNSSPSKESACQETQGAISPIREAEPLPGKDDNPMPPPKSTATVQ 912
S S + P+ S +S +SPS + A++ PL +P PP +A+
Sbjct: 244 PSTSTTQQPKHSPTSPPASPSPSPPSTKPPTAMAS-PSTSPLSATKSHPTPPAAPSASST 420
Query: 913 PSP 915
P P
Sbjct: 421 PPP 429
>TC7897 weakly similar to UP|ENL2_ARATH (Q9T076) Early nodulin-like protein
2 precursor (Phytocyanin-like protein), partial (31%)
Length = 1482
Score = 38.5 bits (88), Expect = 0.006
Identities = 44/170 (25%), Positives = 63/170 (36%), Gaps = 10/170 (5%)
Frame = +3
Query: 755 PKPVASASQKRAKGAGVSTTSGASKLASDAQPEVDDKEGGGDAAIQADVHEHSTSNPPSR 814
PK S S K + +G T+G A P V G S PPS
Sbjct: 546 PKSSPSPSPKASPPSGSPPTAGEPSPAG-TPPSVLPSPAGAPVPAGGP----SAGTPPSA 710
Query: 815 ENSPAPN-----PAKSKAEGVEKPPKAASS----AKKTSTSEGRRKVRSKSGHKSPRRSR 865
SP+P+ PA S A G E P + +S T+TS G +P S
Sbjct: 711 STSPSPSPVSAPPAASPAPGAESPSSSPTSNSPAGGPTATSPSPAGDSPAGGPPAP--SP 884
Query: 866 SSTNSSPSKESACQETQGAISPIREAEPLPGKDDNPMPPP-KSTATVQPS 914
++ ++ PS G S + P +++ PPP ++V PS
Sbjct: 885 TTGDTPPSASGPDVSPSGTPSSLPADTPSSSSNNSTAPPPGNGGSSVVPS 1034
Score = 32.3 bits (72), Expect = 0.45
Identities = 46/188 (24%), Positives = 64/188 (33%), Gaps = 6/188 (3%)
Frame = +3
Query: 788 VDDKEGG---GDAAIQADVHEHSTSNPPS--RENSPAPNPAKSKAEGVEKPPKAASSAKK 842
+ K+G G I + T PPS ++SP+P+P S G PP A
Sbjct: 453 ISGKDGNCEKGQKMILVVISPRGTQPPPSPPPKSSPSPSPKASPPSG--SPPTA------ 608
Query: 843 TSTSEGRRKVRSKSGHKSPRRSRSSTNSSPSKESACQETQGAISPIREAEPLPGKDDNPM 902
G SP + S SP+ A +P P +P
Sbjct: 609 --------------GEPSPAGTPPSVLPSPAGAPVPAGGPSAGTP-------PSASTSPS 725
Query: 903 PPPKST-ATVQPSPQAKGGSSSHVSSEKLDTLFEEDPLAALDGFLDGTLEWDSPPHQADS 961
P P S P+P A+ SSS S+ P A D G PP + +
Sbjct: 726 PSPVSAPPAASPAPGAESPSSSPTSNSPAGGPTATSPSPAGDSPAGG------PPAPSPT 887
Query: 962 TAETAQSS 969
T +T S+
Sbjct: 888 TGDTPPSA 911
>TC15122 similar to UP|Q94AY5 (Q94AY5) At1g19950/T20H2_25, partial (56%)
Length = 1343
Score = 38.1 bits (87), Expect = 0.008
Identities = 31/135 (22%), Positives = 57/135 (41%), Gaps = 9/135 (6%)
Frame = +3
Query: 811 PPSRENSPAPNPAKSKAE---GVEKPPKAASSAKKTSTSEGRRKVRSKSGHKSPRRSRSS 867
P R PAP S A VE+PP SS TS+S+ +++V + G ++ S+
Sbjct: 633 PGVRVRQPAPGKGPSAAATGPNVEEPPSPTSS---TSSSQFQKEVAEELGSPQVPKAAST 803
Query: 868 T----NSSPSKESACQETQGAISPI--REAEPLPGKDDNPMPPPKSTATVQPSPQAKGGS 921
N P + +++ I +++ P P P P ++ +P + +
Sbjct: 804 AAVLNNQKPPVAGGLTTQKSSLAGISTQKSNPAPDTTIKSAPAPAEPKQIEAAPPSSSPA 983
Query: 922 SSHVSSEKLDTLFEE 936
+ + + DT+ EE
Sbjct: 984 NENGNPPTKDTIMEE 1028
>TC9682 similar to UP|Q8VZC0 (Q8VZC0) DTDP-glucose 4-6-dehydratase-like
protein, partial (29%)
Length = 731
Score = 37.7 bits (86), Expect = 0.011
Identities = 31/123 (25%), Positives = 50/123 (40%), Gaps = 1/123 (0%)
Frame = +1
Query: 806 HSTSNPPSRENSPAPNPAKSKAEGVEKPPKAASSAKKTS-TSEGRRKVRSKSGHKSPRRS 864
H+ NPP N+P P+P S V ++SS ++ S + S +P +
Sbjct: 259 HTPPNPP---NTPDPSPDPSTTSSVNSVSSSSSSESSSAPPSSSSNPLSPASALTTPHLT 429
Query: 865 RSSTNSSPSKESACQETQGAISPIREAEPLPGKDDNPMPPPKSTATVQPSPQAKGGSSSH 924
+ ++ PS+ S +T A+SP P A P+ QA G++S
Sbjct: 430 PTPSSPLPSQTSTPPQTASAVSP-------------PESAGDGYALSSPAEQASSGATSL 570
Query: 925 VSS 927
SS
Sbjct: 571 TSS 579
>TC14982 similar to UP|Q9FPI2 (Q9FPI2) At1g17620, partial (32%)
Length = 1036
Score = 37.4 bits (85), Expect = 0.014
Identities = 32/114 (28%), Positives = 41/114 (35%), Gaps = 8/114 (7%)
Frame = +3
Query: 810 NPPSRENSPAPNPAKSKAEGVEKPPKAASSAKKTSTSEGRRKVRSKSGHKSPRRSRSSTN 869
+PP R NS AP + P AA+SA S S S S S+
Sbjct: 102 SPPQRPNSTAPTALLTALNPSTAAPAAAASAPSAS---------G*SSSSSSSSSSSAVL 254
Query: 870 SSPSKESACQETQGAISPIREA--------EPLPGKDDNPMPPPKSTATVQPSP 915
+SPS S T + SP + +P P +P PP T SP
Sbjct: 255 ASPSTSSTAPTTPPSPSPPSNSPTSTSPPPQPSPPNSTSPSPPQTPTRKTSHSP 416
>TC8094 homologue to UP|EFTU_PEA (O24310) Elongation factor Tu, chloroplast
precursor (EF-Tu), partial (60%)
Length = 1092
Score = 37.0 bits (84), Expect = 0.018
Identities = 28/103 (27%), Positives = 45/103 (43%)
Frame = +2
Query: 806 HSTSNPPSRENSPAPNPAKSKAEGVEKPPKAASSAKKTSTSEGRRKVRSKSGHKSPRRSR 865
HS + PPS + P P PA + PP A+SS + + + + + + + P
Sbjct: 212 HS*TPPPSSTSPPPPPPAATALPSPSAPPAASSSGRSPT*TSAQSATSTTA--RPP*PPP 385
Query: 866 SSTNSSPSKESACQETQGAISPIREAEPLPGKDDNPMPPPKST 908
S S PS + + T + P++ A P + PPP ST
Sbjct: 386 SPWRSPPSATAPPRSTTRSTLPLKSA---PAASPS-TPPPSST 502
>AV428128
Length = 415
Score = 36.2 bits (82), Expect = 0.031
Identities = 31/104 (29%), Positives = 41/104 (38%), Gaps = 2/104 (1%)
Frame = +1
Query: 807 STSNPPSRENSPAPNPAKSKAEGVEKPPKAASSAKKTSTSEGRRKVRSKSGHKSPRRSRS 866
S S PP R SP + P + SSA + STS SK+ + R + S
Sbjct: 31 SLSVPPPRLPSPTDQWTRILPLPDPNPKRTLSSATR-STSSATTSPPSKTSPAAARGAPS 207
Query: 867 STNSSPSKESACQETQGAISPIREAEPLPGKDD--NPMPPPKST 908
ST S + S + SP + P P + PPP ST
Sbjct: 208 STRSHAPEPSLSSTSPTTTSPTSPSTPSPSPSSAVSTKPPPSST 339
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.316 0.132 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,351,916
Number of Sequences: 28460
Number of extensions: 315548
Number of successful extensions: 4141
Number of sequences better than 10.0: 247
Number of HSP's better than 10.0 without gapping: 2960
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3624
length of query: 1170
length of database: 4,897,600
effective HSP length: 100
effective length of query: 1070
effective length of database: 2,051,600
effective search space: 2195212000
effective search space used: 2195212000
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Lotus: description of TM0094b.11


