

BLAST2 result
TBLASTN 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
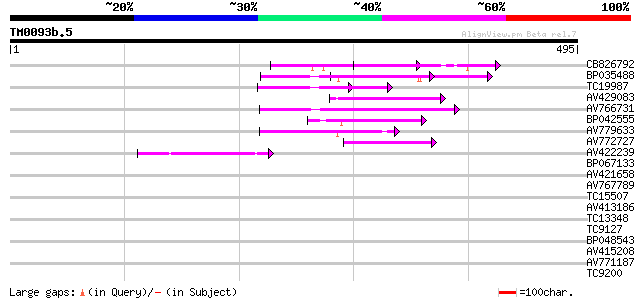
Query= TM0093b.5
(495 letters)
Database: LJGI
28,460 sequences; 14,692,800 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CB826792 59 1e-09
BP035488 56 1e-08
TC19987 weakly similar to UP|Q7XJT3 (Q7XJT3) At2g17140 protein, ... 54 6e-08
AV429083 50 6e-07
AV766731 50 6e-07
BP042555 48 3e-06
AV779633 47 7e-06
AV772727 47 7e-06
AV422239 46 2e-05
BP067133 37 0.010
AV421658 35 0.028
AV767789 34 0.062
TC15507 weakly similar to GB|AAP88326.1|32815835|BT009692 At1g61... 33 0.14
AV413186 32 0.18
TC13348 weakly similar to UP|Q9LUJ4 (Q9LUJ4) Gb|AAF26800.1, part... 32 0.24
TC9127 weakly similar to UP|LYM2_ARATH (O23006) LysM-domain GPI-... 32 0.31
BP048543 32 0.31
AV415208 31 0.41
AV771187 31 0.41
TC9200 similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1, ... 30 0.69
>CB826792
Length = 485
Score = 59.3 bits (142), Expect = 1e-09
Identities = 41/157 (26%), Positives = 74/157 (47%), Gaps = 25/157 (15%)
Frame = +3
Query: 228 RNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERN------------LRHNPSGLV--- 272
R VKEA +++ M+ N V PD+V YN+ ++ LC+ ++ N + +
Sbjct: 15 RLVKEAVNLLQTMEKNNVTPDVVTYNSLIKPLCKNRKIDEAKEVFNDMMKRNITPTIRTF 194
Query: 273 ----------PETLNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGV 322
E ++ +MR PT +Y +L+ + R++ E +I M+ GV
Sbjct: 195 HAFFRILRVEEEVFELLDKMRELGCYPTIETYIMLIRKFCRWRKLDEVFKIWNMMREDGV 374
Query: 323 SPDWVSYYLVVRVLFLSGRFGKGKEIVDQMIGKGLVP 359
S D SY +++ LFL+G+ + + +M KG +P
Sbjct: 375 SHDRSSYIVLIHGLFLNGKVKEAHDYYIEMQRKGFLP 485
Score = 57.8 bits (138), Expect = 4e-09
Identities = 37/130 (28%), Positives = 69/130 (52%), Gaps = 2/130 (1%)
Frame = +3
Query: 301 LGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQMIGKGLVPN 360
L K R VKE+ +L+ M+ + V+PD V+Y +++ L + + + KE+ + M+ + + P
Sbjct: 3 LAKGRLVKEAVNLLQTMEKNNVTPDVVTYNSLIKPLCKNRKIDEAKEVFNDMMKRNITPT 182
Query: 361 HKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPV--YDVLIPKLCRGGDFEKGREL 418
+ + + IL E V EL +KM+ LG Y + Y +LI K CR ++ ++
Sbjct: 183 IRTFHAFFRILRVEEEV---FELLDKMR--ELGCYPTIETYIMLIRKFCRWRKLDEVFKI 347
Query: 419 WDEATSMGIT 428
W+ G++
Sbjct: 348 WNMMREDGVS 377
>BP035488
Length = 534
Score = 56.2 bits (134), Expect = 1e-08
Identities = 42/156 (26%), Positives = 73/156 (45%), Gaps = 4/156 (2%)
Frame = +3
Query: 220 LLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVM 279
+L GW V N EA+R+ K++ ++ PDL Y TF++ L ++ G + L +
Sbjct: 87 ILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKK-------GKLGTALKLF 245
Query: 280 MEMRSH--KVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLF 337
M + P + N ++ L +RV E+ ++ + MK G P+ +Y +++ L
Sbjct: 246 RGMWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVATYNSLIKHLC 425
Query: 338 LSGRFGKGKEIVDQMIGK--GLVPNHKFYFSLIGIL 371
R K E+V+ M K +PN Y L+ L
Sbjct: 426 KIRRMEKVYELVEDMERKKGSCMPNAVTYSCLLNSL 533
Score = 43.9 bits (102), Expect = 6e-05
Identities = 36/143 (25%), Positives = 64/143 (44%), Gaps = 2/143 (1%)
Frame = +3
Query: 281 EMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSG 340
E + H+ + T +N++L+ E+ ++ + + S PD +Y ++ L G
Sbjct: 48 EFQLHRDIKT---WNVILNGWCVLGNAHEAKRVWKDIMASKCRPDLFTYATFIKALTKKG 218
Query: 341 RFGKGKEIVDQMIGKGL--VPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLGGYGPV 398
+ G ++ M +G P+ +I LC +RV ALE+F+ MK
Sbjct: 219 KLGTALKLFRGMWNEGCNCKPDVVICNCIIDALCFKKRVPEALEVFQDMKERGCEPNVAT 398
Query: 399 YDVLIPKLCRGGDFEKGRELWDE 421
Y+ LI LC+ EK EL ++
Sbjct: 399 YNSLIKHLCKIRRMEKVYELVED 467
>TC19987 weakly similar to UP|Q7XJT3 (Q7XJT3) At2g17140 protein, partial
(12%)
Length = 350
Score = 53.9 bits (128), Expect = 6e-08
Identities = 29/118 (24%), Positives = 60/118 (50%)
Frame = +3
Query: 217 YRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETL 276
Y + ++ + + A R++K+M+ NG L YN+ + L + G + E
Sbjct: 9 YDTFIWKFCKEGKISSALRVLKDMERNGCSKTLQTYNSLILGLGSK-------GQIFEMY 167
Query: 277 NVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVR 334
+M EMR + P +YN ++SCL + + +++ +L M G+SP+ S+ ++++
Sbjct: 168 GLMDEMRERGICPDICTYNNVISCLCEGGKTEDATSLLHEMLDKGISPNISSFKILIK 341
Score = 45.1 bits (105), Expect = 3e-05
Identities = 25/85 (29%), Positives = 40/85 (46%), Gaps = 1/85 (1%)
Frame = +3
Query: 217 YRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETL 276
Y SL+ G + E ++ EM+ G+ PD+ YN + CLCE G +
Sbjct: 114 YNSLILGLGSKGQIFEMYGLMDEMRERGICPDICTYNNVISCLCE-------GGKTEDAT 272
Query: 277 NVMMEMRSHKVLPTSISYNILL-SC 300
+++ EM + P S+ IL+ SC
Sbjct: 273 SLLHEMLDKGISPNISSFKILIKSC 347
>AV429083
Length = 316
Score = 50.4 bits (119), Expect = 6e-07
Identities = 32/101 (31%), Positives = 55/101 (53%)
Frame = +3
Query: 280 MEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLS 339
ME++++ + P ++NIL+ L K +VK + +L M GV+PD V+Y ++ L+
Sbjct: 15 MELKNN-IKPDVSTFNILVDALCKKGKVKHAKNVLAVMIKQGVAPDLVTYSTLLDGYCLT 191
Query: 340 GRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHA 380
KGK +++ M G+ PN Y +I C V+ V+ A
Sbjct: 192 KDMYKGKYVLNAMGRVGVTPNVDSYNIVINGFCKVKLVDEA 314
Score = 34.3 bits (77), Expect = 0.048
Identities = 25/81 (30%), Positives = 36/81 (43%)
Frame = +3
Query: 230 VKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLP 289
VK A+ ++ M GV PDLV Y+T L C + V+ M V P
Sbjct: 93 VKHAKNVLAVMIKQGVAPDLVTYSTLLDGYCLTKDMYKGK-------YVLNAMGRVGVTP 251
Query: 290 TSISYNILLSCLGKTRRVKES 310
SYNI+++ K + V E+
Sbjct: 252 NVDSYNIVINGFCKVKLVDEA 314
>AV766731
Length = 576
Score = 50.4 bits (119), Expect = 6e-07
Identities = 36/174 (20%), Positives = 80/174 (45%)
Frame = -3
Query: 219 SLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNV 278
SL+ G R + A ++ M G DL+ N+ LC+ + L+ + + +
Sbjct: 532 SLINGLCKARRISCAVELVDVMHETGPPADLITTNSLFDGLCKHH-------LLDKAIAL 374
Query: 279 MMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFL 338
M+++ K+ P Y +++ L K R+K + +I + + + G + + ++Y +++
Sbjct: 373 FMKVKDPKIQPNIPPYPVIIDGLCKVGRLKIAQEIFQVLLSEGYNLNVMTYTVMINGYCK 194
Query: 339 SGRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSL 392
G + + ++ +M G +PN + ++I L + A LF +M + L
Sbjct: 193 EGLLDEAQALLSKMEDNGCIPNAVNFQTIICALFEKNENDKAERLFREMIARDL 32
>BP042555
Length = 510
Score = 48.1 bits (113), Expect = 3e-06
Identities = 35/113 (30%), Positives = 54/113 (46%), Gaps = 9/113 (7%)
Frame = +3
Query: 261 ERNLRHNPSGLVPETLNVMMEMRSHKVL---------PTSISYNILLSCLGKTRRVKESC 311
ERNL L LNV ++H +L P+ +YN L+ + R KE+
Sbjct: 183 ERNLLSQVHSL----LNVYQVDKAHNMLKYMIVRGFSPSVATYNELIHGYCRVDRFKEAV 350
Query: 312 QILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQMIGKGLVPNHKFY 364
IL M G SPD +YY ++ +G+ GK E+ +M+ KG++P+ Y
Sbjct: 351 GILRDMAKRGXSPDVDTYYPLICGFCQTGKLGKAFEMKAKMVHKGILPDADTY 509
Score = 29.3 bits (64), Expect = 1.5
Identities = 18/78 (23%), Positives = 35/78 (44%)
Frame = +3
Query: 217 YRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETL 276
Y L++G+ KEA I+++M G PD+ Y + C+ +G + +
Sbjct: 297 YNELIHGYCRVDRFKEAVGILRDMAKRGXSPDVDTYYPLICGFCQ-------TGKLGKAF 455
Query: 277 NVMMEMRSHKVLPTSISY 294
+ +M +LP + +Y
Sbjct: 456 EMKAKMVHKGILPDADTY 509
>AV779633
Length = 440
Score = 47.0 bits (110), Expect = 7e-06
Identities = 35/136 (25%), Positives = 58/136 (41%), Gaps = 14/136 (10%)
Frame = -2
Query: 219 SLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNV 278
+L+ G+ + +A +++ GV PD+V YN+ L+ C+ L E L
Sbjct: 400 TLIDGYCEAGLMSQALALMENSWKTGVKPDIVSYNSLLKGFCKAGDLVRAESLFDEILGF 221
Query: 279 MMEMRS--------------HKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSP 324
+ S + PT +Y L+S GK ++ES + E M SG+ P
Sbjct: 220 QRDGESGQLKNNAVDTRDELRNIRPTLATYTTLISAYGKHCGIEESRSLYEQMVMSGIMP 41
Query: 325 DWVSYYLVVRVLFLSG 340
D L+V + F+ G
Sbjct: 40 D----VLLVILFFMEG 5
Score = 27.7 bits (60), Expect = 4.5
Identities = 14/63 (22%), Positives = 32/63 (50%)
Frame = -2
Query: 292 ISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQ 351
I N L+ + + ++ ++E +GV PD VSY +++ +G + + + D+
Sbjct: 412 IGLNTLIDGYCEAGLMSQALALMENSWKTGVKPDIVSYNSLLKGFCKAGDLVRAESLFDE 233
Query: 352 MIG 354
++G
Sbjct: 232 ILG 224
>AV772727
Length = 528
Score = 47.0 bits (110), Expect = 7e-06
Identities = 26/81 (32%), Positives = 42/81 (51%)
Frame = -2
Query: 292 ISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQ 351
I+Y+ LL L K V ++ +LE +K G+ PD +Y +++ L SGR +E+
Sbjct: 524 ITYHSLLDALCKNHHVDKAIALLERVKDKGIQPDMYTYNIIIDGLCKSGRVKDAQELFQD 345
Query: 352 MIGKGLVPNHKFYFSLIGILC 372
++ KG N Y +I LC
Sbjct: 344 LLIKGYRLNVVTYNIMINGLC 282
Score = 35.8 bits (81), Expect = 0.016
Identities = 19/92 (20%), Positives = 48/92 (51%)
Frame = -2
Query: 249 LVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLPTSISYNILLSCLGKTRRVK 308
++ Y++ L LC+ + V + + ++ ++ + P +YNI++ L K+ RVK
Sbjct: 527 VITYHSLLDALCKNHH-------VDKAIALLERVKDKGIQPDMYTYNIIIDGLCKSGRVK 369
Query: 309 ESCQILEAMKTSGVSPDWVSYYLVVRVLFLSG 340
++ ++ + + G + V+Y +++ L + G
Sbjct: 368 DAQELFQDLLIKGYRLNVVTYNIMINGLCIEG 273
Score = 35.8 bits (81), Expect = 0.016
Identities = 25/105 (23%), Positives = 47/105 (43%)
Frame = -2
Query: 181 TVTAIINALCSKGHAKRAEGVVLHHKDKITGALPCIYRSLLYGWSVHRNVKEARRIIKEM 240
T ++++ALC H +A ++ KDK Y ++ G VK+A+ + +++
Sbjct: 521 TYHSLLDALCKNHHVDKAIALLERVKDKGIQPDMYTYNIIIDGLCKSGRVKDAQELFQDL 342
Query: 241 KSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSH 285
G ++V YN + LC GL E L ++++ H
Sbjct: 341 LIKGYRLNVVTYNIMINGLC-------IEGLSDEALALLIQNGRH 228
Score = 35.4 bits (80), Expect = 0.021
Identities = 21/85 (24%), Positives = 41/85 (47%)
Frame = -2
Query: 217 YRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETL 276
Y SLL + +V +A +++ +K G+ PD+ YN + LC+ SG V +
Sbjct: 518 YHSLLDALCKNHHVDKAIALLERVKDKGIQPDMYTYNIIIDGLCK-------SGRVKDAQ 360
Query: 277 NVMMEMRSHKVLPTSISYNILLSCL 301
+ ++ ++YNI+++ L
Sbjct: 359 ELFQDLLIKGYRLNVVTYNIMINGL 285
Score = 32.3 bits (72), Expect = 0.18
Identities = 21/84 (25%), Positives = 40/84 (47%)
Frame = -2
Query: 327 VSYYLVVRVLFLSGRFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEK 386
++Y+ ++ L + K +++++ KG+ P+ Y +I LC RV A ELF+
Sbjct: 524 ITYHSLLDALCKNHHVDKAIALLERVKDKGIQPDMYTYNIIIDGLCKSGRVKDAQELFQD 345
Query: 387 MKSSSLGGYGPVYDVLIPKLCRGG 410
+ Y+++I LC G
Sbjct: 344 LLIKGYRLNVVTYNIMINGLCIEG 273
>AV422239
Length = 387
Score = 45.8 bits (107), Expect = 2e-05
Identities = 29/119 (24%), Positives = 56/119 (46%)
Frame = +2
Query: 112 YNYALRVFAEKQDYTSMDILIGDLKKEGRVMDAQTFGLVAENLIKLGKEDEALGIFKNLD 171
Y+ + A + Y M ++ ++ +G +++ +TF +V + K DEA+ F +D
Sbjct: 29 YHLMIESLARIRQYQIMWDIVTKMRNKG-MLNVETFCIVMRKYARAHKVDEAVYTFNVMD 205
Query: 172 KYKCSTNEFTVTAIINALCSKGHAKRAEGVVLHHKDKITGALPCIYRSLLYGWSVHRNV 230
KY+ N +++ALC + ++A+ + K + L Y LL GW N+
Sbjct: 206 KYEVPQNLAAFNGLLSALCKSRNVRKAQEIFDSMKGRFEPDLK-TYSILLEGWGKDPNL 379
>BP067133
Length = 507
Score = 36.6 bits (83), Expect = 0.010
Identities = 24/112 (21%), Positives = 42/112 (37%), Gaps = 3/112 (2%)
Frame = +3
Query: 155 IKLGKEDEALGIFKNLDKYKCSTNEFTVTAIINALCSKGHAKRAEGVVLHHKDKITGA-- 212
+K G D A+ +F + + C I L H A +H+ +
Sbjct: 171 VKAGLIDNAIKVFDEMTQSNCRLFSIDYNRFIGVLIRHFHLHLAHHY--YHRHVLPNGFS 344
Query: 213 -LPCIYRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCERN 263
LP Y + ++ +++ M S G +PD+ +N +L LC N
Sbjct: 345 LLPFTYSRFISALCAVKDFTSIDTLLQHMDSLGFVPDIWAFNIYLTLLCREN 500
>AV421658
Length = 281
Score = 35.0 bits (79), Expect = 0.028
Identities = 24/90 (26%), Positives = 41/90 (44%)
Frame = +1
Query: 240 MKSNGVIPDLVCYNTFLRCLCERNLRHNPSGLVPETLNVMMEMRSHKVLPTSISYNILLS 299
M + PD+V YNT + C+ P + + + EM +LP + +Y L+
Sbjct: 7 MTERDLSPDVVTYNTLISKFCKLK---EPD--LEKAFEMKAEMVHKGILPDADTYEPLIR 171
Query: 300 CLGKTRRVKESCQILEAMKTSGVSPDWVSY 329
L +R+ E+ + M GVSP+ +Y
Sbjct: 172 TLCLQQRLSEAYDLFREMLRWGVSPNNETY 261
>AV767789
Length = 560
Score = 33.9 bits (76), Expect = 0.062
Identities = 26/102 (25%), Positives = 45/102 (43%)
Frame = -1
Query: 270 GLVPETLNVMMEMRSHKVLPTSISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSY 329
G E +V+ E + PT ++YN+L++ + + + Q+L+ M + PD V+Y
Sbjct: 560 GQYMEARDVISEFGKIGLHPTVMTYNMLMNAYARGGQHSKLPQLLKEMAALNLKPDSVTY 381
Query: 330 YLVVRVLFLSGRFGKGKEIVDQMIGKGLVPNHKFYFSLIGIL 371
++ F + QMI G V + Y L IL
Sbjct: 380 STMIYAFVRVRDFRRAFFYHKQMIKSGQVMDVDSYQKLRAIL 255
>TC15507 weakly similar to GB|AAP88326.1|32815835|BT009692 At1g61870
{Arabidopsis thaliana;}, partial (24%)
Length = 746
Score = 32.7 bits (73), Expect = 0.14
Identities = 18/56 (32%), Positives = 24/56 (42%)
Frame = +3
Query: 371 LCGVERVNHALELFEKMKSSSLGGYGPVYDVLIPKLCRGGDFEKGRELWDEATSMG 426
+ G + A ++F MK P Y L LC GGDFE E+ E+ G
Sbjct: 72 IAGKGTLYEAKKVFADMKKRGYKPESPCYFTLAHFLCEGGDFESAFEITKESMDKG 239
Score = 30.4 bits (67), Expect = 0.69
Identities = 12/30 (40%), Positives = 17/30 (56%)
Frame = +3
Query: 232 EARRIIKEMKSNGVIPDLVCYNTFLRCLCE 261
EA+++ +MK G P+ CY T LCE
Sbjct: 96 EAKKVFADMKKRGYKPESPCYFTLAHFLCE 185
>AV413186
Length = 398
Score = 32.3 bits (72), Expect = 0.18
Identities = 21/81 (25%), Positives = 40/81 (48%), Gaps = 1/81 (1%)
Frame = +1
Query: 292 ISYNILLSCLGKTRRVKESCQILEAMKTSGVSPDWVSYYLVVRVLFLSGRFGKGKEIVDQ 351
+ +N ++S L E+ +L M SGV P+ ++ +++ LSG +G +
Sbjct: 79 VLWNTMISALAHHGFGTEAMVMLNDMIRSGVEPNRATFVVLLNACSLSGLVQEGLQFFKF 258
Query: 352 MIGK-GLVPNHKFYFSLIGIL 371
M + G+VP+ + Y L +L
Sbjct: 259 MTSEFGVVPDQEHYACLTDLL 321
>TC13348 weakly similar to UP|Q9LUJ4 (Q9LUJ4) Gb|AAF26800.1, partial (7%)
Length = 539
Score = 32.0 bits (71), Expect = 0.24
Identities = 13/46 (28%), Positives = 24/46 (51%)
Frame = +3
Query: 217 YRSLLYGWSVHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCER 262
Y +++ H + A R++KEM+ PDL Y+ L+ C++
Sbjct: 45 YNTMISTACAHSREETALRLLKEMEERSCTPDLETYHPLLKMCCKK 182
>TC9127 weakly similar to UP|LYM2_ARATH (O23006) LysM-domain GPI-anchored
protein 2 precursor, partial (46%)
Length = 1016
Score = 31.6 bits (70), Expect = 0.31
Identities = 17/39 (43%), Positives = 23/39 (58%)
Frame = +3
Query: 2 KFKMLNSAWKLCCLRKTRTRNFQLLSASLCSTLQPISAP 40
KFK+LNS W+L C + +R+ SA ST + IS P
Sbjct: 54 KFKLLNSIWRLVCFPLSDSRSITKSSAVSLSTFE-ISTP 167
>BP048543
Length = 547
Score = 31.6 bits (70), Expect = 0.31
Identities = 30/156 (19%), Positives = 59/156 (37%), Gaps = 6/156 (3%)
Frame = +2
Query: 112 YNYALRVFAEKQDYTSMDILIGDLKKEGRVMDAQTFGLVAENLIKLGKEDEALGIFKNLD 171
+N L Y + L L+ +G D T ++ LG+ + A + +
Sbjct: 89 FNNILGSLVRMNHYPTAVSLSQQLELKGIAPDIATLTILINCFCHLGRMNYAFSVLGKIL 268
Query: 172 KYKCSTNEFTVTAIINALCSKGHAKRAEGVVLHHKDKITG------ALPCIYRSLLYGWS 225
K + +T ++ LC G + A + H K G + YR L+
Sbjct: 269 KRAYQPDTTALTTLMKGLCLNGEIRSA--INFHDDVKAKGFQFQVDRVSVTYRFLINELC 442
Query: 226 VHRNVKEARRIIKEMKSNGVIPDLVCYNTFLRCLCE 261
A +++++++ D+ YNT ++ +CE
Sbjct: 443 EVGETGAALQLLRQIEEEHT--DVQMYNTIIKSMCE 544
>AV415208
Length = 242
Score = 31.2 bits (69), Expect = 0.41
Identities = 19/51 (37%), Positives = 24/51 (46%)
Frame = +3
Query: 445 KPVRPEKISLADSPIAKSPKKAKKLLGKVKMRKKPTAVKKKKKRQKKSGAT 495
KP + + +P K PK KK+ K T +KKKKR KKS T
Sbjct: 45 KPAEKKPAAAEKAPAEKKPKAEKKI------SKDATGSEKKKKRTKKSVET 179
>AV771187
Length = 484
Score = 31.2 bits (69), Expect = 0.41
Identities = 20/71 (28%), Positives = 30/71 (42%), Gaps = 1/71 (1%)
Frame = -3
Query: 341 RFGKGKEIVDQMIGKGLVPNHKFYFSLIGILCGVERVNHALELFEKMKSSSLG-GYGPVY 399
R + E+ + M KG PN Y SLI ++ A+E+ M+ + G Y
Sbjct: 239 RIQRALELFEDMKKKGCAPNRVTYDSLIRYYSATNEIDRAVEVLRDMQRLNHGIPSSSSY 60
Query: 400 DVLIPKLCRGG 410
+I LC G
Sbjct: 59 TPIIHALCEAG 27
>TC9200 similar to UP|Q8LLE6 (Q8LLE6) RNA-binding protein AKIP1, partial
(31%)
Length = 727
Score = 30.4 bits (67), Expect = 0.69
Identities = 18/32 (56%), Positives = 21/32 (65%), Gaps = 1/32 (3%)
Frame = +2
Query: 461 KSPKK-AKKLLGKVKMRKKPTAVKKKKKRQKK 491
KS KK KK K K RK+ VKKKKK++KK
Sbjct: 203 KSKKK*RKKWRRKKKRRKRKRRVKKKKKKKKK 298
Database: LJGI
Posted date: Jul 30, 2004 11:16 AM
Number of letters in database: 14,692,800
Number of sequences in database: 28,460
Lambda K H
0.320 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,978,644
Number of Sequences: 28460
Number of extensions: 131001
Number of successful extensions: 798
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 765
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 792
length of query: 495
length of database: 4,897,600
effective HSP length: 94
effective length of query: 401
effective length of database: 2,222,360
effective search space: 891166360
effective search space used: 891166360
frameshift window, decay const: 50, 0.1
T: 13
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 57 (26.6 bits)
Lotus: description of TM0093b.5


